This is a preprint.
Computational Peptide Discovery with a Genetic Programming Approach
- PMID: 37693481
- PMCID: PMC10491332
- DOI: 10.21203/rs.3.rs-3307450/v1
Computational Peptide Discovery with a Genetic Programming Approach
Update in
-
Computational peptide discovery with a genetic programming approach.J Comput Aided Mol Des. 2024 Apr 3;38(1):17. doi: 10.1007/s10822-024-00558-0. J Comput Aided Mol Des. 2024. PMID: 38570405 Free PMC article.
Abstract
Background: The development of peptides for therapeutic targets or biomarkers for disease diagnosis is a challenging task in protein engineering. Current approaches are tedious, often time-consuming and require complex laboratory data due to the vast search space. In silico methods can accelerate research and substantially reduce costs. Evolutionary algorithms are a promising approach for exploring large search spaces and facilitating the discovery of new peptides.
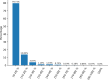
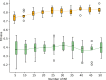
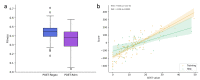
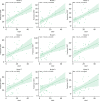
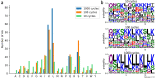
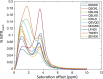
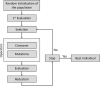
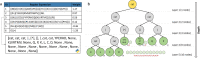
Results: This study presents the development and use of a variant of the initial POET algorithm, called , which is based on genetic programming, where individuals are represented by a list of regular expressions. The program was trained on a small curated dataset and employed to predict new peptides that can improve the problem of sensitivity in detecting peptides through magnetic resonance imaging using chemical exchange saturation transfer (CEST). The resulting model achieves a performance gain of 20% over the initial POET variant and is able to predict a candidate peptide with a 58% performance increase compared to the gold-standard peptide.
Conclusions: By combining the power of genetic programming with the flexibility of regular expressions, new potential peptide targets were identified to improve the sensitivity of detection by CEST. This approach provides a promising research direction for the efficient identification of peptides with therapeutic or diagnostic potential.
Keywords: CEST MRI; Evolutionary algorithm; Genetic programming; Peptide discovery; contrast agent; regular expressions.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures



References
-
- Hökfelt T, Broberger C, Xu ZQD, Sergeyev V, Ubink R, Diez M. Neuropeptides — an overview. Neuropharmacology. 2000;39(8):1337–56. - PubMed
-
- Lj Zhang, Gallo RL. Antimicrobial peptides. Current Biology. 2016;26(1):14–9. - PubMed
-
- Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM. Venoms, venomics, antivenomics. FEBS Letters. 2009;583(11):1736–43. - PubMed
-
- King GF. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opinion on Biological Therapy. 2011;11(11):1469–84. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

