This is a preprint.
Distinct genomic signatures and modifiable risk factors underly the comorbidity between major depressive disorder and cardiovascular disease
- PMID: 37693619
- PMCID: PMC10491387
- DOI: 10.1101/2023.09.01.23294931
Distinct genomic signatures and modifiable risk factors underly the comorbidity between major depressive disorder and cardiovascular disease
Update in
-
Distinct biological signature and modifiable risk factors underlie the comorbidity between major depressive disorder and cardiovascular disease.Nat Cardiovasc Res. 2024 Jun;3(6):754-769. doi: 10.1038/s44161-024-00488-y. Epub 2024 Jun 17. Nat Cardiovasc Res. 2024. PMID: 39215135 Free PMC article.
Abstract
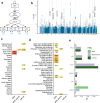
Major depressive disorder (MDD) and cardiovascular disease (CVD) are often comorbid, resulting in excess morbidity and mortality. Using genomic data, this study elucidates biological mechanisms, key risk factors, and causal pathways underlying their comorbidity. We show that CVDs share a large proportion of their genetic risk factors with MDD. Multivariate genome-wide association analysis of the shared genetic liability between MDD and atherosclerotic CVD (ASCVD) revealed seven novel loci and distinct patterns of tissue and brain cell-type enrichments, suggesting a role for the thalamus. Part of the genetic overlap was explained by shared inflammatory, metabolic, and psychosocial/lifestyle risk factors. Finally, we found support for causal effects of genetic liability to MDD on CVD risk, but not from most CVDs to MDD, and demonstrated that the causal effects were partly explained by metabolic and psychosocial/lifestyle factors. The distinct signature of MDD-ASCVD comorbidity aligns with the idea of an immunometabolic sub-type of MDD more strongly associated with CVD than overall MDD. In summary, we identify plausible biological mechanisms underlying MDD-CVD comorbidity, as well as key modifiable risk factors for prevention of CVD in individuals with MDD.
Conflict of interest statement
Competing interests CML sits on the SAB of Myriad Neuroscience and has received speaker/consultancy fees from SYNLAB and UCB. OAA is a consultant to Cortechs.ai, and has received speaker’s honoraria from Lundbeck, Janssen and Sunovion. PFS has receieved consulting fees from and is a shareholder of Neumora therapeutics. Other authors declare no competing interests.
Figures








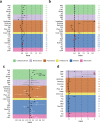






 Indicates that the observed statistically significant association suffered from pleiotropy; possible causal effect should not be interpreted. Psychosocial=Psychosocial/Lifestyle; MDD=Major Depressive Disorder; MDD-ASCVD=common factor for CVD and MDD and ASCVD; PAD=Peripheral Artery Disease; HF=Heart Failure; STR=Stroke; CAD=Coronary Artery Disease; AF=Atrial Fibrillation; DBP=Diastolic Blood Pressure; SBP=Systolic Blood Pressure; PP=Pulse Pressure; Edu=Educational attainment; Phys. Act.=Physical activity; Child. Mal.= Childhood Maltreatment; T2D=Type II Diabetes; TG=Triglycerides; HDL=High-Density Lipoprotein; NonHDL=Non-High-Density Lipoprotein; TC=Total Cholesterol; LDL=Low-Density Lipoprotein; IL6=Interleukin-6; CRP=C-Reactive Protein; MR=Mendelian randomization IVW=Inverse Variance Weighted.
Indicates that the observed statistically significant association suffered from pleiotropy; possible causal effect should not be interpreted. Psychosocial=Psychosocial/Lifestyle; MDD=Major Depressive Disorder; MDD-ASCVD=common factor for CVD and MDD and ASCVD; PAD=Peripheral Artery Disease; HF=Heart Failure; STR=Stroke; CAD=Coronary Artery Disease; AF=Atrial Fibrillation; DBP=Diastolic Blood Pressure; SBP=Systolic Blood Pressure; PP=Pulse Pressure; Edu=Educational attainment; Phys. Act.=Physical activity; Child. Mal.= Childhood Maltreatment; T2D=Type II Diabetes; TG=Triglycerides; HDL=High-Density Lipoprotein; NonHDL=Non-High-Density Lipoprotein; TC=Total Cholesterol; LDL=Low-Density Lipoprotein; IL6=Interleukin-6; CRP=C-Reactive Protein; MR=Mendelian randomization IVW=Inverse Variance Weighted.References
-
- Nemeroff C. B. & Goldschmidt-Clermont P. J. Heartache and heartbreak—the link between depression and cardiovascular disease. Nat Rev Cardiol 9, 526–539 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
