Tangent functional connectomes uncover more unique phenotypic traits
- PMID: 37694156
- PMCID: PMC10483051
- DOI: 10.1016/j.isci.2023.107624
Tangent functional connectomes uncover more unique phenotypic traits
Abstract
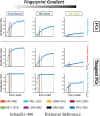
Functional connectomes (FCs) containing pairwise estimations of functional couplings between pairs of brain regions are commonly represented by correlation matrices. As symmetric positive definite matrices, FCs can be transformed via tangent space projections, resulting into tangent-FCs. Tangent-FCs have led to more accurate models predicting brain conditions or aging. Motivated by the fact that tangent-FCs seem to be better biomarkers than FCs, we hypothesized that tangent-FCs have also a higher fingerprint. We explored the effects of six factors: fMRI condition, scan length, parcellation granularity, reference matrix, main-diagonal regularization, and distance metric. Our results showed that identification rates are systematically higher when using tangent-FCs across the "fingerprint gradient" (here including test-retest, monozygotic and dizygotic twins). Highest identification rates were achieved when minimally (0.01) regularizing FCs while performing tangent space projection using Riemann reference matrix and using correlation distance to compare the resulting tangent-FCs. Such configuration was validated in a second dataset (resting-state).
Keywords: Biological sciences; Phenotyping.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Fornito A., Zalesky A., Bullmore E. Academic Press; 2016. Fundamentals of Brain Network Analysis.
-
- Sporns O. MIT Press; 2016. Networks of the Brain.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

