Breakdown of Arabidopsis thaliana thioredoxins and glutaredoxins based on electrostatic similarity-Leads to common and unique interaction partners and functions
- PMID: 37695767
- PMCID: PMC10495010
- DOI: 10.1371/journal.pone.0291272
Breakdown of Arabidopsis thaliana thioredoxins and glutaredoxins based on electrostatic similarity-Leads to common and unique interaction partners and functions
Abstract
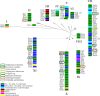
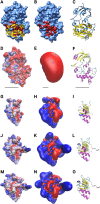
The reversible reduction and oxidation of protein thiols was first described as mechanism to control light/dark-dependent metabolic regulation in photosynthetic organisms. Today, it is recognized as an essential mechanism of regulation and signal transduction in all kingdoms of life. Proteins of the thioredoxin (Trx) family, Trxs and glutaredoxins (Grxs) in particular, catalyze thiol-disulfide exchange reactions and are vital players in the operation of thiol switches. Various Trx and Grx isoforms are present in all compartments of the cell. These proteins have a rather broad but at the same time distinct substrate specificity. Understanding the molecular basis of their target specificity is central to the understanding of physiological and pathological redox signaling. Electrostatic complementarity of the redoxins with their target proteins has been proposed as a major reason. Here, we analyzed the electrostatic similarity of all Arabidopsis thaliana Trxs, Grxs, and proteins containing such domains. Clustering of the redoxins based on this comparison suggests overlapping and also distant target specificities and thus functions of the different sub-classes including all Trx isoforms as well as the three classes of Grxs, i.e. CxxC-, CGFS-, and CC-type Grxs. Our analysis also provides a rationale for the tuned substrate specificities of both the ferredoxin- and NADPH-dependent Trx reductases.
Copyright: © 2023 Bodnar et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Substrate specificity of thioredoxins and glutaredoxins - towards a functional classification.Heliyon. 2019 Dec 17;5(12):e02943. doi: 10.1016/j.heliyon.2019.e02943. eCollection 2019 Dec. Heliyon. 2019. PMID: 31890941 Free PMC article.
-
A fluorescent probe for specifically measuring the overall thioredoxin and glutaredoxin reducing activity in bacterial cells.Analyst. 2022 Feb 28;147(5):834-840. doi: 10.1039/d1an01644j. Analyst. 2022. PMID: 35107099
-
The specificity of thioredoxins and glutaredoxins is determined by electrostatic and geometric complementarity.Chem Sci. 2015 Dec 1;6(12):7049-7058. doi: 10.1039/c5sc01501d. Epub 2015 Sep 9. Chem Sci. 2015. PMID: 29861944 Free PMC article.
-
Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions.Free Radic Res. 2016;50(2):206-45. doi: 10.3109/10715762.2015.1120864. Free Radic Res. 2016. PMID: 26573728 Review.
-
Involvement of thiol-based mechanisms in plant development.Biochim Biophys Acta. 2015 Aug;1850(8):1479-96. doi: 10.1016/j.bbagen.2015.01.023. Epub 2015 Feb 9. Biochim Biophys Acta. 2015. PMID: 25676896 Review.
Cited by
-
Chloroplasts lacking class I glutaredoxins are functional but show a delayed recovery of protein cysteinyl redox state after oxidative challenge.Redox Biol. 2024 Feb;69:103015. doi: 10.1016/j.redox.2023.103015. Epub 2023 Dec 28. Redox Biol. 2024. PMID: 38183796 Free PMC article.
-
Post-Translational Modifications to Cysteine Residues in Plant Proteins and Their Impact on the Regulation of Metabolism and Signal Transduction.Int J Mol Sci. 2024 Sep 12;25(18):9845. doi: 10.3390/ijms25189845. Int J Mol Sci. 2024. PMID: 39337338 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

