OpenABC enables flexible, simplified, and efficient GPU accelerated simulations of biomolecular condensates
- PMID: 37695778
- PMCID: PMC10513381
- DOI: 10.1371/journal.pcbi.1011442
OpenABC enables flexible, simplified, and efficient GPU accelerated simulations of biomolecular condensates
Abstract
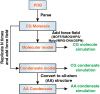
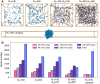
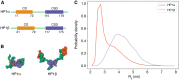
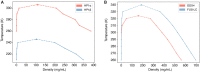
Biomolecular condensates are important structures in various cellular processes but are challenging to study using traditional experimental techniques. In silico simulations with residue-level coarse-grained models strike a balance between computational efficiency and chemical accuracy. They could offer valuable insights by connecting the emergent properties of these complex systems with molecular sequences. However, existing coarse-grained models often lack easy-to-follow tutorials and are implemented in software that is not optimal for condensate simulations. To address these issues, we introduce OpenABC, a software package that greatly simplifies the setup and execution of coarse-grained condensate simulations with multiple force fields using Python scripting. OpenABC seamlessly integrates with the OpenMM molecular dynamics engine, enabling efficient simulations with performance on a single GPU that rivals the speed achieved by hundreds of CPUs. We also provide tools that convert coarse-grained configurations to all-atom structures for atomistic simulations. We anticipate that OpenABC will significantly facilitate the adoption of in silico simulations by a broader community to investigate the structural and dynamical properties of condensates.
Copyright: © 2023 Liu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Update of
-
OpenABC Enables Flexible, Simplified, and Efficient GPU Accelerated Simulations of Biomolecular Condensates.bioRxiv [Preprint]. 2023 Apr 21:2023.04.19.537533. doi: 10.1101/2023.04.19.537533. bioRxiv. 2023. Update in: PLoS Comput Biol. 2023 Sep 11;19(9):e1011442. doi: 10.1371/journal.pcbi.1011442. PMID: 37131742 Free PMC article. Updated. Preprint.
Similar articles
-
OpenABC Enables Flexible, Simplified, and Efficient GPU Accelerated Simulations of Biomolecular Condensates.bioRxiv [Preprint]. 2023 Apr 21:2023.04.19.537533. doi: 10.1101/2023.04.19.537533. bioRxiv. 2023. Update in: PLoS Comput Biol. 2023 Sep 11;19(9):e1011442. doi: 10.1371/journal.pcbi.1011442. PMID: 37131742 Free PMC article. Updated. Preprint.
-
Toward Predictive Coarse-Grained Simulations of Biomolecular Condensates.Biochemistry. 2025 Apr 15;64(8):1750-1761. doi: 10.1021/acs.biochem.4c00737. Epub 2025 Apr 2. Biochemistry. 2025. PMID: 40172489 Review.
-
Protocol for the development of coarse-grained structures for macromolecular simulation using GROMACS.PLoS One. 2023 Aug 3;18(8):e0288264. doi: 10.1371/journal.pone.0288264. eCollection 2023. PLoS One. 2023. PMID: 37535543 Free PMC article.
-
An implementation of the Martini coarse-grained force field in OpenMM.Biophys J. 2023 Jul 25;122(14):2864-2870. doi: 10.1016/j.bpj.2023.04.007. Epub 2023 Apr 11. Biophys J. 2023. PMID: 37050876 Free PMC article.
-
Deciphering driving forces of biomolecular phase separation from simulations.Curr Opin Struct Biol. 2025 Jun;92:103026. doi: 10.1016/j.sbi.2025.103026. Epub 2025 Mar 8. Curr Opin Struct Biol. 2025. PMID: 40058249 Review.
Cited by
-
Cellular Function of a Biomolecular Condensate Is Determined by Its Ultrastructure.bioRxiv [Preprint]. 2024 Dec 27:2024.12.27.630454. doi: 10.1101/2024.12.27.630454. bioRxiv. 2024. PMID: 39763716 Free PMC article. Preprint.
-
Transferable Coarse Graining via Contrastive Learning of Graph Neural Networks.bioRxiv [Preprint]. 2023 Sep 12:2023.09.08.556923. doi: 10.1101/2023.09.08.556923. bioRxiv. 2023. Update in: ACS Cent Sci. 2023 Nov 16;9(12):2286-2297. doi: 10.1021/acscentsci.3c01160. PMID: 37745447 Free PMC article. Updated. Preprint.
-
Chemically Informed Coarse-Graining of Electrostatic Forces in Charge-Rich Biomolecular Condensates.ACS Cent Sci. 2025 Feb 11;11(2):302-321. doi: 10.1021/acscentsci.4c01617. eCollection 2025 Feb 26. ACS Cent Sci. 2025. PMID: 40028356 Free PMC article.
-
Transferable Implicit Solvation via Contrastive Learning of Graph Neural Networks.ACS Cent Sci. 2023 Nov 16;9(12):2286-2297. doi: 10.1021/acscentsci.3c01160. eCollection 2023 Dec 27. ACS Cent Sci. 2023. PMID: 38161379 Free PMC article.
-
Fundamental Aspects of Phase-Separated Biomolecular Condensates.Chem Rev. 2024 Jul 10;124(13):8550-8595. doi: 10.1021/acs.chemrev.4c00138. Epub 2024 Jun 17. Chem Rev. 2024. PMID: 38885177 Free PMC article. Review.

