Transgenic expression of the HERV-W envelope protein leads to polarized glial cell populations and a neurodegenerative environment
- PMID: 37695891
- PMCID: PMC10515160
- DOI: 10.1073/pnas.2308187120
Transgenic expression of the HERV-W envelope protein leads to polarized glial cell populations and a neurodegenerative environment
Abstract
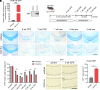
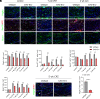
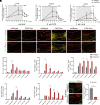
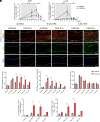
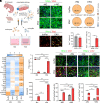
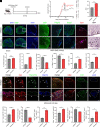
The human endogenous retrovirus type W (HERV-W) has been identified and repeatedly confirmed as human-specific pathogenic entity affecting many cell types in multiple sclerosis (MS). Our recent contributions revealed the encoded envelope (ENV) protein to disturb myelin repair by interfering with oligodendroglial precursor differentiation and by polarizing microglial cells toward an axon-damage phenotype. Indirect proof of ENV's antiregenerative and degenerative activities has been gathered recently in clinical trials using a neutralizing anti-ENV therapeutic antibody. Yet direct proof of its mode of action can only be presented here based on transgenic ENV expression in mice. Upon demyelination, we observed myelin repair deficits, neurotoxic microglia and astroglia, and increased axon degeneration. Experimental autoimmune encephalomyelitis activity progressed faster in mutant mice equally accompanied by activated glial cells. This study therefore provides direct evidence on HERV-W ENV's contribution to the overall negative impact of this activated viral entity in MS.
Keywords: endogenous retrovirus; glia; multiple sclerosis; myelin repair; neurodegeneration.
Conflict of interest statement
P.K. was supported by Stifterverband/Novartisstiftung and received consulting/travel/congress grants from GeNeuro, Sanofi and Servier. H.-P.H. has received fees for consulting, speaking and serving on steering committees from Bayer Healthcare, Biogen, GeNeuro, MedImmune, Merck, Novartis, Opexa, Receptos Celgene, Roche, Sanofi Genzyme, CSL Behring, Octapharma and Teva, with approval from the Rector of HHU. J.G. was supported by Stifterverband/Novartisstiftung and received travel expenses for attending meetings from GeNeuro. B.C. and H.P. receive compensation from GeNeuro-Innovation for their work. All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Perron H., et al. , Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particles. Res. Virol. 140, 551–561 (1989). - PubMed
-
- Dolei A., Perron H., The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: A biological interface between virology, genetics, and immunology in human physiology and disease. J. Neurovirol. 15, 4–13 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

