Mixed, nonclassical behavior in a classic allosteric protein
- PMID: 37695919
- PMCID: PMC10515163
- DOI: 10.1073/pnas.2308338120
Mixed, nonclassical behavior in a classic allosteric protein
Abstract
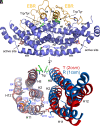
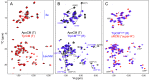
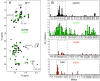
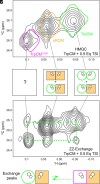
Allostery is a major driver of biological processes requiring coordination. Thus, it is one of the most fundamental and remarkable phenomena in nature, and there is motivation to understand and manipulate it to a multitude of ends. Today, it is often described in terms of two phenomenological models proposed more than a half-century ago involving only T(tense) or R(relaxed) conformations. Here, methyl-based NMR provides extensive detail on a dynamic T to R switch in the classical dimeric allosteric protein, yeast chorismate mutase (CM), that occurs in the absence of substrate, but only with the activator bound. Switching of individual subunits is uncoupled based on direct observation of mixed TR states in the dimer. This unique finding excludes both classic models and solves the paradox of a coexisting hyperbolic binding curve and highly skewed substrate-free T-R equilibrium. Surprisingly, structures of the activator-bound and effector-free forms of CM appear the same by NMR, providing another example of the need to account for dynamic ensembles. The apo enzyme, which has a sigmoidal activity profile, is shown to switch, not to R, but to a related high-energy state. Thus, the conformational repertoire of CM does not just change as a matter of degree depending on the allosteric input, be it effector and/or substrate. Rather, the allosteric model appears to completely change in different contexts, which is only consistent with modern ensemble-based frameworks.
Keywords: Allostery; MWC; NMR dynamics; ensemble allosteric model; relaxation dispersion.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Monod J., Wyman J., Changeux J. P., On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965). - PubMed
-
- Koshland D. E. Jr., Nemethy G., Filmer D., Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 (1966). - PubMed
-
- Schirmer T., Evans P. R., Structural basis of the allosteric behaviour of phosphofructokinase. Nature 343, 140–145 (1990). - PubMed
-
- Stevens R. C., Gouaux J. E., Lipscomb W. N., Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: Crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution. Biochemistry 29, 7691–7701 (1990). - PubMed
-
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B., Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J. Mol. Biol. 204, 725–747 (1988). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

