Mitochondrial membrane potential acts as a retrograde signal to regulate cell cycle progression
- PMID: 37696576
- PMCID: PMC10494934
- DOI: 10.26508/lsa.202302091
Mitochondrial membrane potential acts as a retrograde signal to regulate cell cycle progression
Abstract
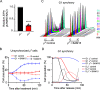
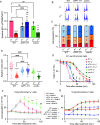
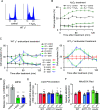
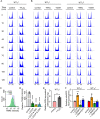
Mitochondria are central to numerous metabolic pathways whereby mitochondrial dysfunction has a profound impact and can manifest in disease. The consequences of mitochondrial dysfunction can be ameliorated by adaptive responses that rely on crosstalk from the mitochondria to the rest of the cell. Such mito-cellular signalling slows cell cycle progression in mitochondrial DNA-deficient (ρ0) Saccharomyces cerevisiae cells, but the initial trigger of the response has not been thoroughly studied. Here, we show that decreased mitochondrial membrane potential (ΔΨm) acts as the initial signal of mitochondrial stress that delays G1-to-S phase transition in both ρ0 and control cells containing mtDNA. Accordingly, experimentally increasing ΔΨm was sufficient to restore timely cell cycle progression in ρ0 cells. In contrast, cellular levels of oxidative stress did not correlate with the G1-to-S delay. Restored G1-to-S transition in ρ0 cells with a recovered ΔΨm is likely attributable to larger cell size, whereas the timing of G1/S transcription remained delayed. The identification of ΔΨm as a regulator of cell cycle progression may have implications for disease states involving mitochondrial dysfunction.
© 2023 Gorospe et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
