A targeted sequencing extension for transcript genotyping in single-cell transcriptomics
- PMID: 37696578
- PMCID: PMC10494938
- DOI: 10.26508/lsa.202301971
A targeted sequencing extension for transcript genotyping in single-cell transcriptomics
Abstract
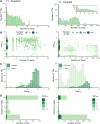
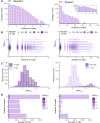
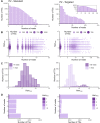
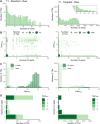
As no existing methods within the single-cell RNA sequencing repertoire combine genotyping of specific genomic loci with high throughput, we evaluated a straightforward, targeted sequencing approach as an extension to high-throughput droplet-based single-cell RNA sequencing. Overlaying standard gene expression data with transcript level genotype information provides a strategy to study the impact of genetic variants. Here, we describe this targeted sequencing extension, explain how to process the data and evaluate how technical parameters such as amount of input cDNA, number of amplification rounds, and sequencing depth influence the number of transcripts detected. Finally, we demonstrate how targeted sequencing can be used in two contexts: (1) simultaneous investigation of the presence of a somatic variant and its potential impact on the transcriptome of affected cells and (2) evaluation of allele-specific expression of a germline variant in ad hoc cell subsets. Through these and other comparable applications, our targeted sequencing extension has the potential to improve our understanding of functional effects caused by genetic variation.
© 2023 Van Horebeek et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





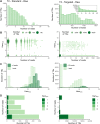


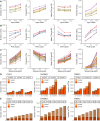






References
-
- Cardamone G, Paraboschi EM, Soldà G, Cantoni C, Supino D, Piccio L, Duga S, Asselta R (2019) Not only cancer: The long non-coding RNA MALAT1 affects the repertoire of alternatively spliced transcripts and circular RNAs in multiple sclerosis. Hum Mol Genet 28: 1414–1428. 10.1093/hmg/ddy438 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
