Dynamics of the DYNLL1-MRE11 complex regulate DNA end resection and recruitment of Shieldin to DSBs
- PMID: 37696958
- PMCID: PMC10686051
- DOI: 10.1038/s41594-023-01074-9
Dynamics of the DYNLL1-MRE11 complex regulate DNA end resection and recruitment of Shieldin to DSBs
Abstract
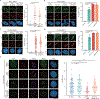
The extent and efficacy of DNA end resection at DNA double-strand breaks (DSB) determine the repair pathway choice. Here we describe how the 53BP1-associated protein DYNLL1 works in tandem with the Shieldin complex to protect DNA ends. DYNLL1 is recruited to DSBs by 53BP1, where it limits end resection by binding and disrupting the MRE11 dimer. The Shieldin complex is recruited to a fraction of 53BP1-positive DSBs hours after DYNLL1, predominantly in G1 cells. Shieldin localization to DSBs depends on MRE11 activity and is regulated by the interaction of DYNLL1 with MRE11. BRCA1-deficient cells rendered resistant to PARP inhibitors by the loss of Shieldin proteins can be resensitized by the constitutive association of DYNLL1 with MRE11. These results define the temporal and functional dynamics of the 53BP1-centric DNA end resection factors in cells.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
A.D.D. reports consulting for AstraZeneca, Bayer AG, Blacksmith/ Lightstone Ventures, Bristol Myers Squibb, Cyteir Therapeutics, EMD Serono, Impact Therapeutics, PrimeFour Therapeutics, Pfizer, Tango Therapeutics and Zentalis Pharmaceuticals/Zeno Management; is an Advisory Board member for Cyteir and Impact Therapeutics; a stockholder in Cedilla Therapeutics, Cyteir, Impact Therapeutics and PrimeFour Therapeutics, and reports receiving commercial research grants from Bristol Myers Squibb, EMD Serono, Moderna and Tango Therapeutics. The remaining authors declare no competing interests.
Figures


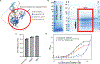












Update of
-
Dynamics of the DYNLL1/MRE11 complex regulates DNA end resection and recruitment of the Shieldin complex to DSBs.bioRxiv [Preprint]. 2023 Mar 27:2023.03.27.534416. doi: 10.1101/2023.03.27.534416. bioRxiv. 2023. Update in: Nat Struct Mol Biol. 2023 Oct;30(10):1456-1467. doi: 10.1038/s41594-023-01074-9. PMID: 37034578 Free PMC article. Updated. Preprint.
References
-
- Hustedt N. & Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

