Integrated profiling identifies CACNG3 as a prognostic biomarker for patients with glioma
- PMID: 37697240
- PMCID: PMC10494363
- DOI: 10.1186/s12885-023-10896-1
Integrated profiling identifies CACNG3 as a prognostic biomarker for patients with glioma
Abstract
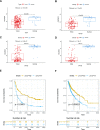
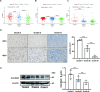
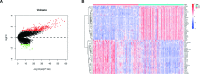
Gliomas are the most common malignant primary brain tumors in adults with poor prognoses. The purpose of this study is to explore CACNG3 as a prognostic factor that is closely related to the progression and survival outcome of gliomas and to provide a potential new molecular target for the diagnosis and treatment of glioma patients. CACNG3 expression and related clinical data were collected from three major databases of The Chinese Glioma Genome Atlas (CGGA), The Cancer Genome Atlas (TCGA), and Gene Expression Omnibus (GEO). The CGGA dataset was used as a training set, and TCGA and GEO datasets obtained from the GEO database were used for validation. CACNG3 was expressed at low levels in the tumor group, and the overall survival (OS) in patients with low CACNG3 expression is shorter. Furthermore, CACNG3 expression was negatively associated with glioma grades, which was confirmed in the IHC results of clinical samples. The expression level of CACNG3 in the IDH1 wide-type group, 1p/19q non-codel group, and mesenchymal subtype group was significantly reduced, and the results showed that CACNG3 could serve as a biomarker for the mesenchymal molecular subtype. In addition, the univariate and multivariate analysis verified the prognostic value of CACNG3 in predicting the OS of gliomas of all grades. The results of functional annotation and pathway enrichment analysis of differently expressed genes(DEGs), showed that CACNG3 might affect the development of glioma by interfering with synaptic transmission. Moreover, temozolomide (TMZ), commonly used in the treatment of glioma, increased CACNG3 expression in a dose and time-dependent manner. Therefore, CACNG3 plays a vital role in the occurrence and development of gliomas and can serve as a potential biomarker for targeted therapy and further investigation in the future.
Keywords: Biomarker; CACNG3; Glioma; Overall survival; Prognostic factor.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

