Diversity of RNA viruses in the cosmopolitan monoxenous trypanosomatid Leptomonas pyrrhocoris
- PMID: 37697369
- PMCID: PMC10496375
- DOI: 10.1186/s12915-023-01687-y
Diversity of RNA viruses in the cosmopolitan monoxenous trypanosomatid Leptomonas pyrrhocoris
Abstract
Background: Trypanosomatids are parasitic flagellates well known because of some representatives infecting humans, domestic animals, and cultural plants. Many trypanosomatid species bear RNA viruses, which, in the case of human pathogens Leishmania spp., influence the course of the disease. One of the close relatives of leishmaniae, Leptomonas pyrrhocoris, has been previously shown to harbor viruses of the groups not documented in other trypanosomatids. At the same time, this species has a worldwide distribution and high prevalence in the natural populations of its cosmopolitan firebug host. It therefore represents an attractive model to study the diversity of RNA viruses.
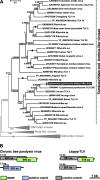
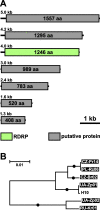
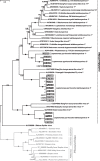
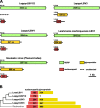
Results: We surveyed 106 axenic cultures of L. pyrrhocoris and found that 64 (60%) of these displayed 2-12 double-stranded RNA fragments. The analysis of next-generation sequencing data revealed four viral groups with seven species, of which up to five were simultaneously detected in a single trypanosomatid isolate. Only two of these species, a tombus-like virus and an Ostravirus, were earlier documented in L. pyrrhocoris. In addition, there were four new species of Leishbuviridae, the family encompassing trypanosomatid-specific viruses, and a new species of Qinviridae, the family previously known only from metatranscriptomes of invertebrates. Currently, this is the only qinvirus with an unambiguously determined host. Our phylogenetic inferences suggest reassortment in the tombus-like virus owing to the interaction of different trypanosomatid strains. Two of the new Leishbuviridae members branch early on the phylogenetic tree of this family and display intermediate stages of genomic segment reduction between insect Phenuiviridae and crown Leishbuviridae.
Conclusions: The unprecedented wide range of viruses in one protist species and the simultaneous presence of up to five viral species in a single Leptomonas pyrrhocoris isolate indicate the uniqueness of this flagellate. This is likely determined by the peculiarity of its firebug host, a highly abundant cosmopolitan species with several habits ensuring wide distribution and profuseness of L. pyrrhocoris, as well as its exposure to a wider spectrum of viruses compared to other trypanosomatids combined with a limited ability to transmit these viruses to its relatives. Thus, L. pyrrhocoris represents a suitable model to study the adoption of new viruses and their relationships with a protist host.
Keywords: Leishbuviridae; Ostravirus; Pyrrhocoris apterus; Qinviridae; Tombus-like viruses.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Maslov DA, Opperdoes FR, Kostygov AY, Hashimi H, Lukeš J, Yurchenko V. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology. 2019;146(1):1–27. - PubMed
-
- Kostygov AY, Yurchenko V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae) Folia Parasitol. 2017;64:020. - PubMed
-
- Votýpka J, Klepetková H, Yurchenko VY, Horák A, Lukeš J, Maslov DA. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist. 2012;163(4):616–631. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

