Combination of RUNX1 inhibitor and gemcitabine mitigates chemo-resistance in pancreatic ductal adenocarcinoma by modulating BiP/PERK/eIF2α-axis-mediated endoplasmic reticulum stress
- PMID: 37697370
- PMCID: PMC10494371
- DOI: 10.1186/s13046-023-02814-x
Combination of RUNX1 inhibitor and gemcitabine mitigates chemo-resistance in pancreatic ductal adenocarcinoma by modulating BiP/PERK/eIF2α-axis-mediated endoplasmic reticulum stress
Abstract
Background: Gemcitabine (GEM)-based chemotherapy is the first-line option for pancreatic ductal adenocarcinoma (PDAC). However, the development of drug resistance limits its efficacy, and the specific mechanisms remain largely unknown. RUNX1, a key transcription factor in hematopoiesis, also involved in the malignant progression of PDAC, but was unclear in the chemoresistance of PDAC.
Methods: Comparative analysis was performed to screen GEM-resistance related genes using our single-cell RNA sequencing(scRNA-seq) data and two public RNA-sequencing datasets (GSE223463, GSE183795) for PDAC. The expression of RUNX1 in PDAC tissues was detected by qRT-PCR, immunohistochemistry (IHC) and western blot. The clinical significance of RUNX1 in PDAC was determined by single-or multivariate analysis and survival analysis. We constructed the stably expressing cell lines with shRUNX1 and RUNX1, and successfully established GEM-resistant cell line. The role of RUNX1 in GEM resistance was determined by CCK8 assay, plate colony formation assay and apoptosis analysis in vitro and in vivo. To explore the mechanism, we performed bioinformatic analysis using the scRNA-seq data to screen for the endoplasm reticulum (ER) stress signaling that was indispensable for RUNX1 in GEM resistance. We observed the cell morphology in ER stress by transmission electron microscopy and validated RUNX1 in gemcitabine resistance depended on the BiP/PERK/eIF2α pathway by in vitro and in vivo oncogenic experiments, using ER stress inhibitor(4-PBA) and PERK inhibitor (GSK2606414). The correlation between RUNX1 and BiP expression was assessed using the scRNA-seq data and TCGA dataset, and validated by RT-PCR, immunostaining and western blot. The mechanism of RUNX1 regulation of BiP was confirmed by ChIP-PCR and dual luciferase assay. Finally, the effect of RUNX1 inhibitor on PDAC was conducted in vivo mouse models, including subcutaneous xenograft and patient-derived xenograft (PDX) mouse models.
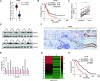
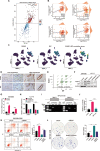
Results: RUNX1 was aberrant high expressed in PDAC and closely associated with GEM resistance. Silencing of RUNX1 could attenuate resistance in GEM-resistant cell line, and its inhibitor Ro5-3335 displayed an enhanced effect in inhibiting tumor growth, combined with GEM treatment, in PDX mouse models and GEM-resistant xenografts. In detail, forced expression of RUNX1 in PDAC cells suppressed apoptosis induced by GEM exposure, which was reversed by the ER stress inhibitor 4-PBA and PERK phosphorylation inhibitor GSK2606414. RUNX1 modulation of ER stress signaling mediated GEM resistance was supported by the analysis of scRNA-seq data. Consistently, silencing of RUNX1 strongly inhibited the GEM-induced activation of BiP and PERK/eIF2α signaling, one of the major pathways involved in ER stress. It was identified that RUNX1 directly bound to the promoter region of BiP, a primary ER stress sensor, and stimulated BiP expression to enhance the reserve capacity for cell adaptation, which in turn facilitated GEM resistance in PDAC cells.
Conclusions: This study identifies RUNX1 as a predictive biomarker for response to GEM-based chemotherapy. RUNX1 inhibition may represent an effective strategy for overcoming GEM resistance in PDAC cells.
Keywords: BiP; ER stress; Gemcitabine resistance; PDAC; RUNX1.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
The authors have declared no potential conflicts of interest.
Figures





References
-
- Soreide K, Rangelova E, Dopazo C, Mieog S, Stattner S. Pancreatic cancer. Eur J Surg Oncol. 2023;49(2):521–525. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

