Prefoldin 2 contributes to mitochondrial morphology and function
- PMID: 37697385
- PMCID: PMC10496292
- DOI: 10.1186/s12915-023-01695-y
Prefoldin 2 contributes to mitochondrial morphology and function
Abstract
Background: Prefoldin is an evolutionarily conserved co-chaperone of the tailless complex polypeptide 1 ring complex (TRiC)/chaperonin containing tailless complex 1 (CCT). The prefoldin complex consists of six subunits that are known to transfer newly produced cytoskeletal proteins to TRiC/CCT for folding polypeptides. Prefoldin function was recently linked to the maintenance of protein homeostasis, suggesting a more general function of the co-chaperone during cellular stress conditions. Prefoldin acts in an adenosine triphosphate (ATP)-independent manner, making it a suitable candidate to operate during stress conditions, such as mitochondrial dysfunction. Mitochondrial function depends on the production of mitochondrial proteins in the cytosol. Mechanisms that sustain cytosolic protein homeostasis are vital for the quality control of proteins destined for the organelle and such mechanisms among others include chaperones.
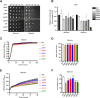
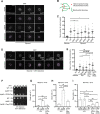
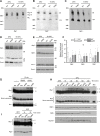
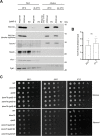
Results: We analyzed consequences of the loss of prefoldin subunits on the cell proliferation and survival of Saccharomyces cerevisiae upon exposure to various cellular stress conditions. We found that prefoldin subunits support cell growth under heat stress. Moreover, prefoldin facilitates the growth of cells under respiratory growth conditions. We showed that mitochondrial morphology and abundance of some respiratory chain complexes was supported by the prefoldin 2 (Pfd2/Gim4) subunit. We also found that Pfd2 interacts with Tom70, a receptor of mitochondrial precursor proteins that are targeted into mitochondria.
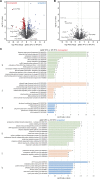
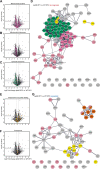
Conclusions: Our findings link the cytosolic prefoldin complex to mitochondrial function. Loss of the prefoldin complex subunit Pfd2 results in adaptive cellular responses on the proteome level under physiological conditions suggesting a continuous need of Pfd2 for maintenance of cellular homeostasis. Within this framework, Pfd2 might support mitochondrial function directly as part of the cytosolic quality control system of mitochondrial proteins or indirectly as a component of the protein homeostasis network.
Keywords: Chaperone; Mitochondria; Pfd2/Gim4; Prefoldin; Proteostasis; Tom70.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

