Hormone Analogues with Unique Signaling Profiles from Replacement of α-Residue Triads with β/γ Diads
- PMID: 37697685
- PMCID: PMC10588032
- DOI: 10.1021/jacs.3c06703
Hormone Analogues with Unique Signaling Profiles from Replacement of α-Residue Triads with β/γ Diads
Abstract
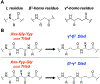
We have applied an underexplored backbone modification strategy to generate new analogues of peptides that activate two clinically important class B1 G protein-coupled receptors (GPCRs). Most peptide modification strategies involve changing side chains or, less commonly, changing the configuration at side chain-bearing carbons (i.e., l residues replaced by d residues). In contrast, backbone modifications alter the number of backbone atoms and the identities of backbone atoms relative to a poly-α-amino acid backbone. Starting from the peptide agonists PTH(1-34) (the first 34 residues of the parathyroid hormone, used clinically as the drug teriparatide) and glucagon-like peptide-1 (7-36) (GLP-1(7-36)), we replaced native α-residue triads with a diad composed of a β-amino acid residue and a γ-amino acid residue. The β/γ diad retains the number of backbone atoms in the ααα triad. Because the β and γ residue each bear a single side chain, we implemented ααα→βγ replacements at sites that contained a Gly residue (i.e., at α-residue triads that presented only two side chains). All seven of the α/β/γ-peptides derived from PTH(1-34) or GLP-1(7-36) bind to the cognate receptor (the PTHR1 or the GLP-1R), but they vary considerably in their activity profiles. Outcomes include functional mimicry of the all-α agonist, receptor-selective agonist activity, biased agonism, or strong binding with weak activation, which could lead to antagonist development. Collectively, these findings demonstrate that ααα→βγ replacements, which are easily implemented via solid-phase synthesis, can generate peptide hormone analogues that display unique and potentially useful signaling behavior.
Figures

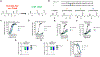

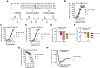

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

