Increased STAT3 Activation in Periodontitis Drives Inflammatory Bone Loss
- PMID: 37697911
- PMCID: PMC10714379
- DOI: 10.1177/00220345231192381
Increased STAT3 Activation in Periodontitis Drives Inflammatory Bone Loss
Abstract
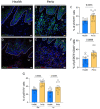
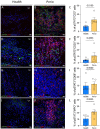
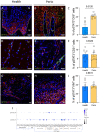
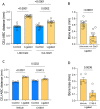
Periodontitis is one of the most prevalent human inflammatory diseases. It is characterized by periodontal tissue destruction, progressively driven by the host response. In this regard, cytokines associated with tissue destruction, such as interleukin (IL)-6 and IL-23, use a common signaling pathway mediated by STAT3. This transcription factor is also needed for IL-17A production, a key mediator in periodontitis pathogenesis. Although several studies have reported increased activation of STAT3 in experimental periodontitis, a detailed characterization of STAT3 activation in human gingival tissues and its involvement in alveolar bone loss has yet to be explored. Using a cross-sectional study design, we detected increased proportions of pSTAT3-positive cells during periodontitis compared with health, particularly in epithelial cells and T cells. Other cell types of hematopoietic and nonhematopoietic origin also display STAT3 activation in gingival tissues. We detected increased STAT3 phosphorylation and expression of STAT3-related genes during experimental periodontitis. Next, we evaluated the role of STAT3 in alveolar bone destruction using a mouse model of STAT3 loss of function (mut-Stat3 mice). Compared with controls, mut-Stat3 mice had reduced alveolar bone loss following ligature-induced periodontitis. We also evaluated pharmacologic inhibition of STAT3 in ligature-induced periodontitis. Like mut-Stat3 mice, mice treated with STAT3 small-molecule inhibitor had reduced bone loss compared with controls. Our results demonstrate that STAT3 activation is increased in epithelial and T cells during periodontitis and indicate a pathogenic role of STAT3 in inflammatory alveolar bone loss.
Keywords: gingiva; inflammation; mouth mucosa; mucosal immunity; periodontal diseases; signal transduction.
Conflict of interest statement
Declaration of Conflicting InterestsThe authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
-
- Abusleme L, Hoare A, Hong BY, Diaz PI. 2021. Microbial signatures of health, gingivitis, and periodontitis. Periodontol 2000. 86(1):57–78. - PubMed
-
- Ambili R, Janam P, Saneesh Babu PS, Prasad M, Vinod D, Anil Kumar PR, Kumary TV, Asha Nair S. 2017. Differential expression of transcription factors NF-κB and STAT3 in periodontal ligament fibroblasts and gingiva of healthy and diseased individuals. Arch Oral Biol. 82:19–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

