Novel multidrug-resistant sublineages of Staphylococcus aureus clonal complex 22 discovered in India
- PMID: 37698417
- PMCID: PMC10597471
- DOI: 10.1128/msphere.00185-23
Novel multidrug-resistant sublineages of Staphylococcus aureus clonal complex 22 discovered in India
Abstract
Staphylococcus aureus is a major pathogen in India causing community and nosocomial infections, but little is known about its molecular epidemiology and mechanisms of resistance in hospital settings. Here, we use whole-genome sequencing (WGS) to characterize 478 S. aureus clinical isolates (393 methicillin-resistant Staphylococcus aureus (MRSA) and 85 methicilin-sensitive Staphylococcus aureus (MSSA) collected from 17 sentinel sites across India between 2014 and 2019. Sequencing results confirmed that sequence type 22 (ST22) (142 isolates, 29.7%), ST239 (74 isolates, 15.48%), and ST772 (67 isolates, 14%) were the most common clones. An in-depth analysis of 175 clonal complex (CC) 22 Indian isolates identified two novel ST22 MRSA lineages, both Panton-Valentine leukocidin+, both resistant to fluoroquinolones and aminoglycosides, and one harboring the the gene for toxic shock syndrome toxin 1 (tst). A temporal analysis of 1797 CC22 global isolates from 14 different studies showed that the two Indian ST22 lineages shared a common ancestor in 1984 (95% highest posterior density [HPD]: 1982-1986), as well as evidence of transmission to other parts of the world. Moreover, the study also gives a comprehensive view of ST2371, a sublineage of CC22, as a new emerging lineage in India and describes it in relationship with the other Indian ST22 isolates. In addition, the retrospective identification of a putative outbreak of multidrug-resistant (MDR) ST239 from a single hospital in Bangalore that persisted over a period of 3 years highlights the need for the implementation of routine surveillance and simple infection prevention and control measures to reduce these outbreaks. To our knowledge, this is the first WGS study that characterized CC22 in India and showed that the Indian clones are distinct from the EMRSA-15 clone. Thus, with the improved resolution afforded by WGS, this study substantially contributed to our understanding of the global population of MRSA. IMPORTANCE The study conducted in India between 2014 and 2019 presents novel insights into the prevalence of MRSA in the region. Previous studies have characterized two dominant clones of MRSA in India, ST772 and ST239, using whole-genome sequencing. However, this study is the first to describe the third dominant clone, ST22, using the same approach. The ST22 Indian isolates were analyzed in-depth, leading to the discovery of two new sublineages of hospital-acquired Staphylococcus aureus in India, both carrying antimicrobial resistance genes and mutations, which limit treatment options for patients. One of the newly characterized sublineages, second Indian cluster, carries the tsst-1 virulence gene, increasing the risk of severe infections. The geographic spread of the two novel lineages, both within India and internationally, could pose a global public health threat. The study also sheds light on ST2371 in India, a single-locus variant of ST22. The identification of a putative outbreak of MDR ST239 in a single hospital in Bangalore emphasizes the need for routine surveillance and simple infection prevention and control measures to reduce these outbreaks. Overall, this study significantly contributes to our understanding of the global population of MRSA, thanks to the improved resolution afforded by WGS.
Keywords: India; ST22; ST239; ST772; Staphylococcus aureus; WGS; outbreak.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
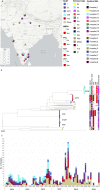
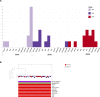

References
-
- WHO publishes list of bacteria for which new antibiotics are urgently needed. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-.... Retrieved . 4 June 2022
-
- Tchamba CN, Touzain F, Fergestad M, De Visscher A, L’Abee-Lund T, De Vliegher S, Wasteson Y, Blanchard Y, Argudín MA, Mainil J, Thiry D. 2023. Identification of staphylococcal cassette chromosome MEC in Staphylococcus aureus and non-aureus staphylococci from dairy cattle in Belgium: comparison of multiplex PCR and whole genome sequencing. Res Vet Sci 155:150–155. doi:10.1016/j.rvsc.2023.01.011 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
