Characterization of a FourU RNA Thermometer in the 5' Untranslated Region of Autolysin Gene blyA in the Bacillus subtilis 168 Prophage SPβ
- PMID: 37699513
- PMCID: PMC10586365
- DOI: 10.1021/acs.biochem.3c00368
Characterization of a FourU RNA Thermometer in the 5' Untranslated Region of Autolysin Gene blyA in the Bacillus subtilis 168 Prophage SPβ
Abstract
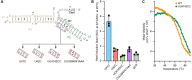
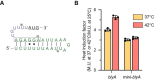
RNA thermometers are noncoding RNA structures located in the 5' untranslated regions (UTRs) of genes that regulate gene expression through temperature-dependent conformational changes. The fourU class of RNA thermometers contains a specific motif in which four consecutive uracil nucleotides are predicted to base pair with the Shine-Dalgarno (SD) sequence in a stem. We employed a bioinformatic search to discover a fourU RNA thermometer in the 5'-UTR of the blyA gene of the Bacillus subtilis phage SPβc2, a bacteriophage that infects B. subtilis 168. blyA encodes an autolysin enzyme, N-acetylmuramoyl-l-alanine amidase, which is involved in the lytic life cycle of the SPβ prophage. We have biochemically validated the predicted RNA thermometer in the 5'-UTR of the blyA gene. Our study suggests that RNA thermometers may play an underappreciated yet critical role in the lytic life cycle of bacteriophages.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
The N-acetylmuramoyl-L-alanine amidase encoded by the Bacillus subtilis 168 prophage SP beta.Microbiology (Reading). 1998 Apr;144 ( Pt 4):885-893. doi: 10.1099/00221287-144-4-885. Microbiology (Reading). 1998. PMID: 9579063
-
Structural and functional characterization of MrpR, the master repressor of the Bacillus subtilis prophage SPβ.Nucleic Acids Res. 2023 Sep 22;51(17):9452-9474. doi: 10.1093/nar/gkad675. Nucleic Acids Res. 2023. PMID: 37602373 Free PMC article.
-
Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis.PLoS Genet. 2014 Oct 9;10(10):e1004636. doi: 10.1371/journal.pgen.1004636. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25299644 Free PMC article.
-
The life cycle of SPβ and related phages.Arch Virol. 2021 Aug;166(8):2119-2130. doi: 10.1007/s00705-021-05116-9. Epub 2021 Jun 7. Arch Virol. 2021. PMID: 34100162 Free PMC article. Review.
-
Closely related and yet special - how SPβ family phages control lysis-lysogeny decisions.Trends Microbiol. 2025 Apr;33(4):387-396. doi: 10.1016/j.tim.2024.11.007. Epub 2024 Dec 6. Trends Microbiol. 2025. PMID: 39645480 Review.
Cited by
-
Robo-Therm, a pipeline to RNA thermometer discovery and validation.RNA. 2024 Jun 17;30(7):760-769. doi: 10.1261/rna.079980.124. RNA. 2024. PMID: 38565243 Free PMC article.
-
Two temperature-responsive RNAs act in concert: the small RNA CyaR and the mRNA ompX.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf041. doi: 10.1093/nar/gkaf041. Nucleic Acids Res. 2025. PMID: 39907110 Free PMC article.
References
-
- Murphy E. R.; Roßmanith J.; Sieg J.; Fris M. E.; Hussein H.; Kouse A. B.; Gross K.; Zeng C.; Hines J. V.; Narberhaus F.; Coschigano P. W. Regulation of ompA translation and Shigella dysenteriae virulence by an RNA thermometer. Infect. Immun. 2020, 88 (3), e00871-19.10.1128/IAI.00871-19. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

