Multi-feature clustering of CTCF binding creates robustness for loop extrusion blocking and Topologically Associating Domain boundaries
- PMID: 37699887
- PMCID: PMC10497529
- DOI: 10.1038/s41467-023-41265-y
Multi-feature clustering of CTCF binding creates robustness for loop extrusion blocking and Topologically Associating Domain boundaries
Abstract
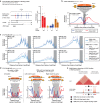
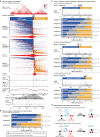
Topologically Associating Domains (TADs) separate vertebrate genomes into insulated regulatory neighborhoods that focus genome-associated processes. TADs are formed by Cohesin-mediated loop extrusion, with many TAD boundaries consisting of clustered binding sites of the CTCF insulator protein. Here we determine how this clustering of CTCF binding contributes to the blocking of loop extrusion and the insulation between TADs. We identify enrichment of three features of CTCF binding at strong TAD boundaries, consisting of strongly bound and closely spaced CTCF binding peaks, with a further enrichment of DNA-binding motifs within these peaks. Using multi-contact Nano-C analysis in cells with normal and perturbed CTCF binding, we establish that individual CTCF binding sites contribute to the blocking of loop extrusion, but in an incomplete manner. When clustered, individual CTCF binding sites thus create a stepwise insulation between neighboring TADs. Based on these results, we propose a model whereby multiple instances of temporal loop extrusion blocking create strong insulation between TADs.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Pushing the TAD boundary: Decoding insulator codes of clustered CTCF sites in 3D genomes.Bioessays. 2024 Oct;46(10):e2400121. doi: 10.1002/bies.202400121. Epub 2024 Aug 21. Bioessays. 2024. PMID: 39169755 Review.
-
Insulation between adjacent TADs is controlled by the width of their boundaries through distinct mechanisms.Proc Natl Acad Sci U S A. 2025 Mar 18;122(11):e2413112122. doi: 10.1073/pnas.2413112122. Epub 2025 Mar 10. Proc Natl Acad Sci U S A. 2025. PMID: 40063813
-
Permeable TAD boundaries and their impact on genome-associated functions.Bioessays. 2024 Oct;46(10):e2400137. doi: 10.1002/bies.202400137. Epub 2024 Aug 2. Bioessays. 2024. PMID: 39093600
-
Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains.Genome Biol. 2020 Jan 7;21(1):5. doi: 10.1186/s13059-019-1894-x. Genome Biol. 2020. PMID: 31910870 Free PMC article.
-
TADs and Their Borders: Free Movement or Building a Wall?J Mol Biol. 2020 Feb 7;432(3):643-652. doi: 10.1016/j.jmb.2019.11.025. Epub 2019 Dec 27. J Mol Biol. 2020. PMID: 31887284 Review.
Cited by
-
Enhancer reprogramming: critical roles in cancer and promising therapeutic strategies.Cell Death Discov. 2025 Mar 3;11(1):84. doi: 10.1038/s41420-025-02366-3. Cell Death Discov. 2025. PMID: 40032852 Free PMC article. Review.
-
Outward-oriented sites within clustered CTCF boundaries are key for intra-TAD chromatin interactions and gene regulation.Nat Commun. 2023 Dec 7;14(1):8101. doi: 10.1038/s41467-023-43849-0. Nat Commun. 2023. PMID: 38062010 Free PMC article.
-
Interpreting the CTCF-mediated sequence grammar of genome folding with AkitaV2.PLoS Comput Biol. 2025 Feb 4;21(2):e1012824. doi: 10.1371/journal.pcbi.1012824. eCollection 2025 Feb. PLoS Comput Biol. 2025. PMID: 39903776 Free PMC article.
-
Structural perturbation of chromatin domains with multiple developmental regulators can severely impact gene regulation and development.bioRxiv [Preprint]. 2024 Aug 3:2024.08.03.606480. doi: 10.1101/2024.08.03.606480. bioRxiv. 2024. Update in: Dev Cell. 2025 Jul 7;60(13):1838-1853.e9. doi: 10.1016/j.devcel.2025.02.002. PMID: 39372737 Free PMC article. Updated. Preprint.
-
Single-cell Micro-C profiles 3D genome structures at high resolution and characterizes multi-enhancer hubs.Nat Genet. 2025 Jul;57(7):1777-1786. doi: 10.1038/s41588-025-02247-6. Epub 2025 Jul 2. Nat Genet. 2025. PMID: 40603633 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

