Clinical Efficiency of Metagenomic Next-Generation Sequencing in Sputum for Pathogen Detection of Patients with Pneumonia According to Disease Severity and Host Immune Status
- PMID: 37700802
- PMCID: PMC10493106
- DOI: 10.2147/IDR.S419892
Clinical Efficiency of Metagenomic Next-Generation Sequencing in Sputum for Pathogen Detection of Patients with Pneumonia According to Disease Severity and Host Immune Status
Abstract
Purpose: Severe pneumonia causes the highest mortality rate in immunocompromised patients. This study aimed to investigate the pathogen diagnostic efficacy of metagenomic next-generation sequencing (mNGS) using sputum sample in patients with pneumonia according to patients' disease severity and immune conditions.
Patients and methods: A total of 180 patients suffering from pneumonia were recruited, and sputum samples were collected in duplicate for pathogen detection by both conventional microbiological tests (CMT) and mNGS. Then, the performance of pathogen identification was examined between two methods, according to disease severity and patients' immune status.
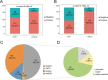
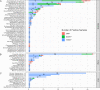
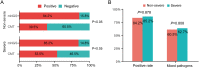
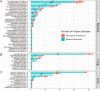
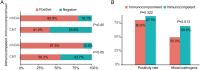
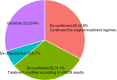
Results: In comparison to CMT, mNGS had higher positivity rates in all patients with pneumonia (85.0% vs 62.2%, P=9.445e-07). The most commonly detected microorganism in sputum of pneumonia patients was Acinetobacter baumannii (42/180, 23.3%) in bacterum level, Candida albicans in fungus level (44/180, 24.4%), and Human herpesvirus 1 (39/180, 27.5%) in virus level. However, for mNGS results, Candida albicans in 34.9% of positive patients, and Human herpesvirus 1 in 7.7% of positive cases were confirmed as pathogens causing pneumonia. Acinetobacter baumannii detected by mNGS in 75% of positive patients was diagnosed as pathogen of pneumonia. The microorganism profile of sputum mNGS differed according to disease severity and immune status of patients. Pneumocystis jirovecii was more likely to infect immunocompromised patients (P=0.002). Pseudomonas aeruginosa (14.8% vs 0.0%, P=0.008) and Human herpesvirus 1 (26.1% vs 5.3%, P=0.004) had higher infection rate in patients with severe pneumonia compared with non-severe cases. mNGS had overwhelming advantages over CMT in detecting a lot of microorganisms including Streptococcus pneumoniae, Enterococcus faecium, Pneumocystis jirovecii, and majority of viruses.
Conclusion: mNGS is a complementary tool of CMT for detecting suspected pathogens for patients with lower respiratory infections. The interpretation of opportunistic pathogens identified by mNGS is challenging, and needs comprehensive consideration of sequencing data and clinical factors.
Keywords: conventional microbiological test; immunocompromised patient; lower respiratory infections; metagenomic-next generation sequencing; severe pneumonia.
© 2023 Chang et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
References
-
- Collaborators GL. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

