Mining the nanotube-forming Bacillus amyloliquefaciens MR14M3 genome for determining anti- Candida auris and anti- Candida albicans potential by pathogenicity and comparative genomics analysis
- PMID: 37701018
- PMCID: PMC10493893
- DOI: 10.1016/j.csbj.2023.08.031
Mining the nanotube-forming Bacillus amyloliquefaciens MR14M3 genome for determining anti- Candida auris and anti- Candida albicans potential by pathogenicity and comparative genomics analysis
Abstract
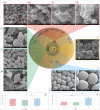
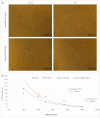
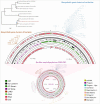
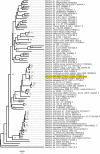
There is a global health concern associated with the emergence of the multidrug-resistant (MDR) fungus Candida auris, which has significant mortality rates. Finding innovative and distinctive anti-Candida compounds is essential for treating infections caused by MDR C. auris. A bacterial strain with anti-Candida activity was isolated and identified using 16 S rRNA gene sequencing. The whole genome was sequenced to identify biosynthesis-related gene clusters. The pathogenicity and cytotoxicity of the isolate were analyzed in Candida and HFF-1 cell lines, respectively. This study set out to show that whole-genome sequencing, cytotoxicity testing, and pathogenicity analysis combined with genome mining and comparative genomics can successfully identify biosynthesis-related gene clusters in native bacterial isolates that encode antifungal natural compounds active against Candida albicans and C. auris. The native isolate MR14M3 has the ability to inhibit C. auris (zone of inhibition 25 mm) and C. albicans (zone of inhibition 25 mm). The 16 S rRNA gene sequence of MR14M3 aligned with Bacillus amyloliquefaciens with similarity (100%). Bacillus amyloliquefaciens MR14M3 establishes bridges of intercellular nanotubes (L 258.56 ± 35.83 nm; W 25.32 ± 6.09 nm) connecting neighboring cells. Candida cell size was reduced significantly, and crushed phenotypes were observed upon treatment with the defused metabolites of B. amyloliquefaciens MR14M3. Furthermore, the pathogenicity of B. amyloliquefaciens MR14M3 on Candida cells was observed through cell membrane disruption and lysed yeast cells. The whole-genome alignment of the MR14M3 genome (3981,643 bp) using 100 genes confirmed its affiliation with Bacillus amyloliquefaciens. Genome mining analysis revealed that MR14M3-coded secondary metabolites are involved in the biosynthesis of polyketides (PKs) and nonribosomal peptide synthases (NRPSs), including 11 biosynthesis-related gene clusters with one hundred percent similarity. Highly conserved biosynthesis-related gene clusters with anti-C. albicans and anti-C. auris potentials and cytotoxic-free activity of B. amyloliquefaciens MR14M3 proposes the utilization of Bacillus amyloliquefaciens MR14M3 as a biofactory for an anti-Candida auris and anti-C. albicans compound synthesizer.
Keywords: Antifungal activity; Bacillus; Biofactory; Biosynthesis-related gene clusters; Candida albicans; Candida auris; Comparative genomics; Cytotoxicity; Genome mining.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Diosgenin producing Bacillus sp. strain IRMC27M2 as a genome-mined weapon against multidrug-resistant Candidozyma (Candida) auris.Comput Struct Biotechnol J. 2025 Jul 30;27:3410-3432. doi: 10.1016/j.csbj.2025.07.048. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40799907 Free PMC article.
-
Genome-Guided Identification of Surfactin-Producing Bacillus halotolerans AQ11M9 with Anti-Candida auris Potential.Int J Mol Sci. 2024 Sep 27;25(19):10408. doi: 10.3390/ijms251910408. Int J Mol Sci. 2024. PMID: 39408762 Free PMC article.
-
Polyketide-derived macrobrevins from marine macroalga-associated Bacillus amyloliquefaciens as promising antibacterial agents against pathogens causing nosocomial infections.Phytochemistry. 2022 Jan;193:112983. doi: 10.1016/j.phytochem.2021.112983. Epub 2021 Oct 23. Phytochemistry. 2022. PMID: 34695706
-
Candida auris: What Have We Learned About Its Mechanisms of Pathogenicity?Front Microbiol. 2018 Dec 12;9:3081. doi: 10.3389/fmicb.2018.03081. eCollection 2018. Front Microbiol. 2018. PMID: 30631313 Free PMC article. Review.
-
Candida auris-the growing menace to global health.Mycoses. 2019 Aug;62(8):620-637. doi: 10.1111/myc.12904. Epub 2019 Jun 18. Mycoses. 2019. PMID: 30773703 Review.
Cited by
-
Diosgenin producing Bacillus sp. strain IRMC27M2 as a genome-mined weapon against multidrug-resistant Candidozyma (Candida) auris.Comput Struct Biotechnol J. 2025 Jul 30;27:3410-3432. doi: 10.1016/j.csbj.2025.07.048. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40799907 Free PMC article.
-
Hybrid Genome and Clinical Impact of Emerging Extensively Drug-Resistant Priority Bacterial Pathogen Acinetobacter baumannii in Saudi Arabia.Life (Basel). 2025 Jul 12;15(7):1094. doi: 10.3390/life15071094. Life (Basel). 2025. PMID: 40724597 Free PMC article.
-
Comparative Analysis of Signature Sequences from Adenylation Domains Situated within Bacterial-Origin Nonribosomal Peptide Synthetase Modules.J Microbiol Biotechnol. 2025 Jul 14;35:e2502030. doi: 10.4014/jmb.2503.02030. J Microbiol Biotechnol. 2025. PMID: 40659553 Free PMC article.
-
Genome-Guided Identification of Surfactin-Producing Bacillus halotolerans AQ11M9 with Anti-Candida auris Potential.Int J Mol Sci. 2024 Sep 27;25(19):10408. doi: 10.3390/ijms251910408. Int J Mol Sci. 2024. PMID: 39408762 Free PMC article.
-
Genomic Landscape of Multidrug Resistance and Virulence in Enterococcus faecalis IRMC827A from a Long-Term Patient.Biology (Basel). 2023 Sep 29;12(10):1296. doi: 10.3390/biology12101296. Biology (Basel). 2023. PMID: 37887006 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
