Resistance to anti-PD-1/anti-PD-L1: galectin-3 inhibition with GB1211 reverses galectin-3-induced blockade of pembrolizumab and atezolizumab binding to PD-1/PD-L1
- PMID: 37701441
- PMCID: PMC10493609
- DOI: 10.3389/fimmu.2023.1250559
Resistance to anti-PD-1/anti-PD-L1: galectin-3 inhibition with GB1211 reverses galectin-3-induced blockade of pembrolizumab and atezolizumab binding to PD-1/PD-L1
Abstract
Background: Galectin-3 (Gal-3) is a β-galactoside-binding lectin that is highly expressed within the tumor microenvironment of aggressive cancers and has been suggested to predict a poor response to immune checkpoint therapy with the anti-PD-1 monoclonal antibody pembrolizumab. We aimed to assess if the effect of Gal-3 was a result of direct interaction with the immune checkpoint receptor.
Methods: The ability of Gal-3 to interact with the PD-1/PD-L1 complex in the absence and presence of blocking antibodies was assessed in in vitro biochemical and cellular assays as well as in an in vivo syngeneic mouse cancer model.
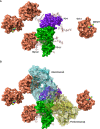
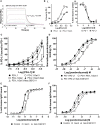
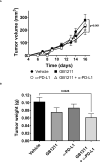
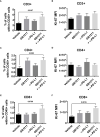
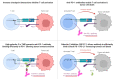
Results: Gal-3 reduced the binding of the checkpoint inhibitors pembrolizumab (anti-PD-1) and atezolizumab (anti-PD-L1), by potentiating the interaction between the PD-1/PD-L1 complex. In the presence of a highly selective Gal-3 small molecule inhibitor (GB1211) the binding of the anti-PD-1/anti-PD-L1 therapeutics was restored to control levels. This was observed in both a surface plasmon resonance assay measuring protein-protein interactions and via flow cytometry. Combination therapy with GB1211 and an anti-PD-L1 blocking antibody reduced tumor growth in an in vivo syngeneic model and increased the percentage of tumor infiltrating T lymphocytes.
Conclusion: Our study suggests that Gal-3 can potentiate the PD-1/PD-L1 immune axis and potentially contribute to the immunosuppressive signalling mechanisms within the tumor microenvironment. In addition, Gal-3 prevents atezolizumab and pembrolizumab target engagement with their respective immune checkpoint receptors. Reversal of this effect with the clinical candidate GB1211 offers a potential enhancing combination therapeutic with anti-PD-1 and -PD-L1 blocking antibodies.
Keywords: PD-1 - PDL-1 axis; atezolizumab; binding; computational chemistry; galectin-3 (Gal3); immuno-oncology; pembrolizumab.
Copyright © 2023 Mabbitt, Holyer, Roper, Nilsson, Zetterberg, Vuong, Mackinnon, Pedersen and Slack.
Conflict of interest statement
UN, FZ, RS, JR, IH, and AM have ownership interest including stock, patents, etc. in Galecto Biotech AB. AP is the COO at Galecto Biotech and has ownership interest including stock, patents, etc. in Galecto Biotech. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Gal-3 blocks the binding between PD-1 and pembrolizumab.J Immunother Cancer. 2024 Oct 2;12(10):e009952. doi: 10.1136/jitc-2024-009952. J Immunother Cancer. 2024. PMID: 39357979 Free PMC article.
-
Inhibition of galectin-3 augments the antitumor efficacy of PD-L1 blockade in non-small-cell lung cancer.FEBS Open Bio. 2021 Mar;11(3):911-920. doi: 10.1002/2211-5463.13088. Epub 2021 Jan 31. FEBS Open Bio. 2021. PMID: 33455075 Free PMC article.
-
Enhancing clinical and immunological effects of anti-PD-1 with belapectin, a galectin-3 inhibitor.J Immunother Cancer. 2021 Apr;9(4):e002371. doi: 10.1136/jitc-2021-002371. J Immunother Cancer. 2021. PMID: 33837055 Free PMC article. Clinical Trial.
-
Study and analysis of antitumor resistance mechanism of PD1/PD-L1 immune checkpoint blocker.Cancer Med. 2020 Nov;9(21):8086-8121. doi: 10.1002/cam4.3410. Epub 2020 Sep 2. Cancer Med. 2020. PMID: 32875727 Free PMC article. Review.
-
The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma.Clin Ther. 2015 Apr 1;37(4):764-82. doi: 10.1016/j.clinthera.2015.02.018. Epub 2015 Mar 29. Clin Ther. 2015. PMID: 25823918 Free PMC article. Review.
Cited by
-
Galectins as pivotal components in oncogenesis and immune exclusion in human malignancies.Front Immunol. 2023 Feb 3;14:1145268. doi: 10.3389/fimmu.2023.1145268. eCollection 2023. Front Immunol. 2023. PMID: 36817445 Free PMC article. Review.
-
Susceptibility of Melanoma Cells to Targeted Therapy Correlates with Protection by Blood Neutrophils.Cancers (Basel). 2024 May 2;16(9):1767. doi: 10.3390/cancers16091767. Cancers (Basel). 2024. PMID: 38730718 Free PMC article.
-
Gal-3 blocks the binding between PD-1 and pembrolizumab.J Immunother Cancer. 2024 Oct 2;12(10):e009952. doi: 10.1136/jitc-2024-009952. J Immunother Cancer. 2024. PMID: 39357979 Free PMC article.
-
Differential Gene Expression of Checkpoint Markers and Cancer Markers in Mouse Models of Spontaneous Chronic Colitis.Cancers (Basel). 2023 Sep 29;15(19):4793. doi: 10.3390/cancers15194793. Cancers (Basel). 2023. PMID: 37835487 Free PMC article.
-
N-Acetylgalactosamine-4-sulfatase (Arylsulfatase B) Regulates PD-L1 Expression in Melanoma by an HDAC3-Mediated Epigenetic Mechanism.Int J Mol Sci. 2024 May 28;25(11):5851. doi: 10.3390/ijms25115851. Int J Mol Sci. 2024. PMID: 38892038 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

