Loss of ATG4B and ATG4A results in two-stage cell cycle defects in pancreatic ductal adenocarcinoma cells
- PMID: 37701987
- PMCID: PMC10617609
- DOI: 10.1242/jcs.260644
Loss of ATG4B and ATG4A results in two-stage cell cycle defects in pancreatic ductal adenocarcinoma cells
Abstract
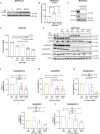
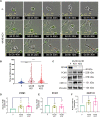
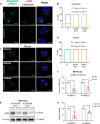
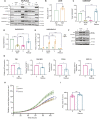
Pancreatic ductal adenocarcinoma (PDAC) exhibits elevated levels of autophagy, which promote tumor progression and treatment resistance. ATG4B is an autophagy-related cysteine protease under consideration as a potential therapeutic target, but it is largely unexplored in PDAC. Here, we investigated the clinical and functional relevance of ATG4B expression in PDAC. Using two PDAC patient cohorts, we found that low ATG4B mRNA or protein expression is associated with worse patient survival outcomes, poorly differentiated PDAC tumors and a lack of survival benefit from adjuvant chemotherapy. In PDAC cell lines, ATG4B knockout reduced proliferation, abolished processing of LC3B (also known as MAP1LC3B), and reduced GABARAP and GABARAPL1 levels, but increased ATG4A levels. ATG4B and ATG4A double knockout lines displayed a further reduction in proliferation, characterized by delays in G1-S phase transition and mitosis. Pro-LC3B accumulated aberrantly at the centrosome with a concomitant increase in centrosomal proteins PCM1 and CEP131, which was rescued by exogenous ATG4B. The two-stage cell cycle defects following ATG4B and ATG4A loss have important therapeutic implications for PDAC.
Keywords: ATG4A; ATG4B; Autophagy; CEP131; Centrosome; Doryphagy; GABARAP; PCM1; PDAC; Pancreatic cancer; pro-LC3B.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Bortnik, S., Choutka, C., Horlings, H. M., Leung, S., Baker, J. H. E., Lebovitz, C., Dragowska, W. H., Go, N. E., Bally, M. B., Minchinton, A. I.et al. (2016). Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 7, 66970-66988. 10.18632/oncotarget.11408 - DOI - PMC - PubMed
-
- Bortnik, S., Tessier-Cloutier, B., Leung, S., Xu, J., Asleh, K., Burugu, S., Magrill, J., Greening, K., Derakhshan, F., Yip, S.et al. (2020). Differential expression and prognostic relevance of autophagy-related markers ATG4B, GABARAP, and LC3B in breast cancer. Breast Cancer Res. Treat. 183, 525-547. 10.1007/s10549-020-05795-z - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

