Structural and antigenic characterization of the avian adeno-associated virus capsid
- PMID: 37702486
- PMCID: PMC10617571
- DOI: 10.1128/jvi.00780-23
Structural and antigenic characterization of the avian adeno-associated virus capsid
Abstract
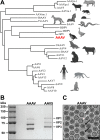
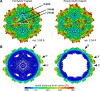
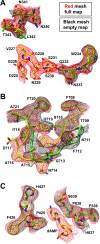
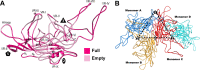
AAVs are extensively studied as promising therapeutic gene delivery vectors. In order to circumvent pre-existing antibodies targeting primate-based AAV capsids, the AAAV capsid was evaluated as an alternative to primate-based therapeutic vectors. Despite the high sequence diversity, the AAAV capsid was found to bind to a common glycan receptor, terminal galactose, which is also utilized by other AAVs already being utilized in gene therapy trials. However, contrary to the initial hypothesis, AAAV was recognized by approximately 30% of human sera tested. Structural and sequence comparisons point to conserved epitopes in the fivefold region of the capsid as the reason determinant for the observed cross-reactivity.
Keywords: AAV vector; adeno-associated virus; antigenicity; avian viruses; capsid; cryo-EM; gene therapy; parvovirus; three-dimensional structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.J Virol. 2022 Jun 8;96(11):e0033522. doi: 10.1128/jvi.00335-22. Epub 2022 May 9. J Virol. 2022. PMID: 35532224 Free PMC article.
-
High-Resolution Structural Characterization of a New Adeno-associated Virus Serotype 5 Antibody Epitope toward Engineering Antibody-Resistant Recombinant Gene Delivery Vectors.J Virol. 2018 Dec 10;93(1):e01394-18. doi: 10.1128/JVI.01394-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30333169 Free PMC article.
-
Structural Study of Aavrh.10 Receptor and Antibody Interactions.J Virol. 2021 Nov 9;95(23):e0124921. doi: 10.1128/JVI.01249-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34549984 Free PMC article.
-
The role of the adeno-associated virus capsid in gene transfer.Methods Mol Biol. 2008;437:51-91. doi: 10.1007/978-1-59745-210-6_2. Methods Mol Biol. 2008. PMID: 18369962 Free PMC article. Review.
-
Parvovirus Capsid-Antibody Complex Structures Reveal Conservation of Antigenic Epitopes Across the Family.Viral Immunol. 2021 Jan-Feb;34(1):3-17. doi: 10.1089/vim.2020.0022. Epub 2020 Apr 21. Viral Immunol. 2021. PMID: 32315582 Free PMC article. Review.
Cited by
-
AAV Vectors Pseudotyped with Capsids from Porcine and Bovine Species Mediate In Vitro and In Vivo Gene Delivery.Viruses. 2023 Dec 29;16(1):57. doi: 10.3390/v16010057. Viruses. 2023. PMID: 38257756 Free PMC article.
-
Genomic loss of GPR108 disrupts AAV transduction in birds.bioRxiv [Preprint]. 2024 May 17:2024.05.16.589954. doi: 10.1101/2024.05.16.589954. bioRxiv. 2024. PMID: 38798475 Free PMC article. Preprint.
-
Birds of a feather flock together: structural characterization of red-crowned crane and turkey aveparvoviruses.J Virol. 2025 Jul 22;99(7):e0011025. doi: 10.1128/jvi.00110-25. Epub 2025 Jul 3. J Virol. 2025. PMID: 40607800 Free PMC article.
-
Adeno-associated viral vector targeted evolution for neurofibromatosis gene delivery.Trends Mol Med. 2025 Apr;31(4):388-398. doi: 10.1016/j.molmed.2025.01.004. Epub 2025 Jan 30. Trends Mol Med. 2025. PMID: 39890493 Review.
-
Structural characterization of antibody-responses following Zolgensma treatment for AAV capsid engineering to expand patient cohorts.Nat Commun. 2025 Apr 19;16(1):3731. doi: 10.1038/s41467-025-59088-4. Nat Commun. 2025. PMID: 40253479 Free PMC article.
References
-
- Microbiology Society . ICTV virus taxonomy profile: parvoviridae. : https://www.microbiologyresearch.org/content/journal/jgv/10.1099/jgv.0.0.... Accessed 16 February 2023 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

