Dissemination of IncI plasmid encoding bla CTX-M-1 is not hampered by its fitness cost in the pig's gut
- PMID: 37702541
- PMCID: PMC10583664
- DOI: 10.1128/aac.00111-23
Dissemination of IncI plasmid encoding bla CTX-M-1 is not hampered by its fitness cost in the pig's gut
Abstract
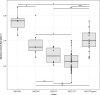
Multiresistance plasmids belonging to the IncI incompatibility group have become one of the most pervasive plasmid types in extended-spectrum beta-lactamase-producing Escherichia coli of animal origin. The extent of the burden imposed on the bacterial cell by these plasmids seems to modulate the emergence of "epidemic" plasmids. However, in vivo data in the natural environment of the strains are scarce. Here, we investigated the cost of a bla CTX-M-1-IncI1 epidemic plasmid in a commensal E. coli animal strain, UB12-RC, before and after oral inoculation of 15 6- to 8-week- old specific-pathogen-free pigs. Growth rate in rich medium was determined on (i) UB12-RC and derivatives, with or without plasmid, in vivo and/or in vitro evolved, and (ii) strains that acquired the plasmid in the gut during the experiment. Although bla CTX-M-1-IncI1 plasmid imposed no measurable burden on the recipient strain after conjugation and during the longitudinal carriage in the pig's gut, we observed a significant difference in the bacterial growth rate between IncI1 plasmid-carrying and plasmid-free isolates collected during in vivo carriage. Only a few mutations on the chromosome of the UB12-RC derivatives were detected by whole-genome sequencing. RNA-Seq analysis of a selected set of these strains showed that transcriptional responses to the bla CTX-M-1-IncI1 acquisition were limited, affecting metabolism, stress response, and motility functions. Our data suggest that the effect of IncI plasmid on host cells is limited, fitness cost being insufficient to act as a barrier to IncI plasmid spread among natural population of E. coli in the gut niche.
Keywords: CTX-M; ESBL; IncI plasmid; RNA-Seq; fitness cost.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Achalapong S, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Babin F-X, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Berkley JA, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Clotaire Donatien R, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Duong Bich T, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garcia C, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Hsia Y, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Jenney AWJ, Khorana M, Khusuwan S, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Lim K, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Manoharan A, Marks F, May J, Mayxay M, Mturi N, Munera-Huertas T, Musicha P, Musila LA, Mussi-Pinhata MM, Naidu RN, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Ochoa TJ, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Ounchanum P, Pak GD, Paredes JL, Peleg AY, Perrone C, Phe T, Phommasone K, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Rattanavong S, Riddell A, Roberts T, Robotham JV, Roca A, Rosenthal VD, Rudd KE, Russell N, Sader HS, Saengchan W, Schnall J, Scott JAG, Seekaew S, Sharland M, Shivamallappa M, Sifuentes-Osornio J, Simpson AJ, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Tigoi C, Turner C, Turner P, van Doorn HR, Velaphi S, Vongpradith A, Vongsouvath M, Vu H, Walsh T, Walson JL, Waner S, Wangrangsimakul T, Wannapinij P, Wozniak T, Young Sharma TEMW, Yu KC, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 - DOI - PMC - PubMed
-
- Guyomard-Rabenirina S, Reynaud Y, Pot M, Albina E, Couvin D, Ducat C, Gruel G, Ferdinand S, Legreneur P, Le Hello S, Malpote E, Sadikalay S, Talarmin A, Breurec S. 2020. Antimicrobial resistance in wildlife in guadeloupe (French West Indies): distribution of a single blaCTX–M–1/IncI1/ST3 plasmid among humans and wild animals. Front Microbiol 11:1524. doi: 10.3389/fmicb.2020.01524 - DOI - PMC - PubMed
-
- Petrin S, Orsini M, Mastrorilli E, Longo A, Cozza D, Olsen JE, Ricci A, Losasso C, Barco L. 2021. Identification and characterization of a spreadable Inci1 Plasmid Harbouring a blaCTX-M-15 gene in an Italian human isolate of Salmonella serovar Napoli. Plasmid 114:102566. doi: 10.1016/j.plasmid.2021.102566 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

