CK1δ and CK1ε Signaling Sustains Mitochondrial Metabolism and Cell Survival in Multiple Myeloma
- PMID: 37702657
- PMCID: PMC10690099
- DOI: 10.1158/0008-5472.CAN-22-2350
CK1δ and CK1ε Signaling Sustains Mitochondrial Metabolism and Cell Survival in Multiple Myeloma
Abstract
Multiple myeloma remains an incurable malignancy due to acquisition of intrinsic programs that drive therapy resistance. Here we report that casein kinase-1δ (CK1δ) and CK1ε are therapeutic targets in multiple myeloma that are necessary to sustain mitochondrial metabolism. Specifically, the dual CK1δ/CK1ε inhibitor SR-3029 had potent in vivo and ex vivo anti-multiple myeloma activity, including against primary multiple myeloma patient specimens. RNA sequencing (RNA-seq) and metabolic analyses revealed inhibiting CK1δ/CK1ε disables multiple myeloma metabolism by suppressing genes involved in oxidative phosphorylation (OxPhos), reducing citric acid cycle intermediates, and suppressing complexes I and IV of the electron transport chain. Finally, sensitivity of multiple myeloma patient specimens to SR-3029 correlated with elevated expression of mitochondrial genes, and RNA-seq from 687 multiple myeloma patient samples revealed that increased CSNK1D, CSNK1E, and OxPhos genes correlate with disease progression and inferior outcomes. Thus, increases in mitochondrial metabolism are a hallmark of multiple myeloma progression that can be disabled by targeting CK1δ/CK1ε.
Significance: CK1δ and CK1ε are attractive therapeutic targets in multiple myeloma whose expression increases with disease progression and connote poor outcomes, and that are necessary to sustain expression of genes directing OxPhos.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures
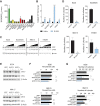
![Figure 1. Pharmacoproteomic screen to identify novel antimyeloma kinase targets. A, Experimental design of activity-based protein profiling of three multiple myeloma cell lines (H929, OPM2, and MM.1S) in the absence or presence of cocultured HS-5 stromal cells after 24 hours. B, Venn diagram illustrating overlap of kinases whose activity is significantly altered by coculture of multiple myeloma cells with stromal cells. C, Heat map depicting the fold change in the activity of ABPP target proteins in each of the three multiple myeloma cell lines [note that CK1δ and CK1ε (red) proteins could not be differentiated by peptide sequence in ABPP]. Log2 ratio of coculture to monoculture is presented. D, Bar graphs of sensitivity of H929, MM.1S, and OPM2 cell lines (EC50) cultured on stroma to a panel of protein kinase inhibitors (at 96 hours).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/82e2/10690099/b4a3213a0a03/3901fig1.gif)
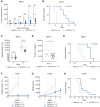
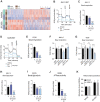
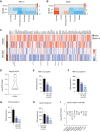
![Figure 4. Inhibition of CK1δ/CK1ε disrupts central metabolic pathways in multiple myeloma patient samples. A, Ex vivo sensitivity of patient CD138+-selected cells to SR-3029 versus a panel of standard-of-care anti–multiple myeloma therapies, as measured by live-cell imaging. Each dot represents sensitivity [lethal dose (LD)50, amount of drug needed to cause lethality in 50% of patient samples] for a given patient sample after 96 hours of treatment. Only patients with measurable LD50 are graphed. Ex vivo responses (achieved LD50 at 96 hours) were as follows: SR-3029 74/75; bortezomib (BTZ) 271/273; carfilzomib (CFZ) 259/263; lenalidomide [Len0 4/203; pomalidomide (Pom) 10/288; dexamethasone (Dex) 17/216; melphalan (Mel) 216/240]. B and C, Ex vivo sensitivity of patient CD138+-selected multiple myeloma cells to SR-3029 versus a panel of kinase inhibitors after 96 hours of treatment (B). Number of patient samples that achieved an ex vivo response (LD50 at 96 hours) out of the total number of samples tested (C) is shown. D, Ex vivo sensitivity of patients to SR-3029 based on clinical course. NDMM prior to therapy (n = 26); early RRMM, early relapse refractory multiple myeloma patients resistant to 1–3 lines of therapy (n = 31); late RRMM, late relapse refractory resistant to ≥4 lines of therapy (n = 34). Statistical comparisons between clinical course were calculated using a one-way ANOVA with multiple comparisons. E–G, CD138+-derived multiple myeloma cells from 5 patients were cultured in the top well Boyden chamber with patient BM-derived stroma in the bottom well. Cells were incubated for 24 hours with vehicle or 250 nmol/L SR-3029 and cells were harvested for RNA-seq analyses. E, Heat map of 1,162 differently expressed genes from the patient RNA-seq data. F, GSEA HALLMARK pathway analysis (q < 0.01) of genes downregulated by SR-3029. G, DAVID pathway analysis of genes downregulated by SR-3029 ex vivo.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/82e2/10690099/2cad04ebb562/3901fig4.gif)
References
-
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med 2011;364:1046–60. - PubMed
-
- Durie BGM, Hoering A, Abidi MH, Rajkumar SV, Epstein J, Kahanic SP, et al. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): a randomised, open-label, phase 3 trial. Lancet 2017;389:519–27. - PMC - PubMed
-
- Dimopoulos MA, Moreau P, Palumbo A, Joshua D, Pour L, Hajek R, et al. Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): a randomised, phase 3, open-label, multicentre study. Lancet Oncol 2016;17:27–38. - PubMed
-
- San-Miguel JF, Hungria VT, Yoon SS, Beksac M, Dimopoulos MA, Elghandour A, et al. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): a randomised, placebo-controlled, phase 3 trial. Lancet Haematol 2016;3:e506–e15. - PubMed
-
- Chen C, Siegel D, Gutierrez M, Jacoby M, Hofmeister CC, Gabrail N, et al. Safety and efficacy of selinexor in relapsed or refractory multiple myeloma and Waldenstrom macroglobulinemia. Blood 2018;131:855–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

