TGF-β Promotes the Postselection Thymic Development and Peripheral Function of IFN-γ-Producing Invariant NKT cells
- PMID: 37702745
- PMCID: PMC10592054
- DOI: 10.4049/jimmunol.2200809
TGF-β Promotes the Postselection Thymic Development and Peripheral Function of IFN-γ-Producing Invariant NKT cells
Abstract
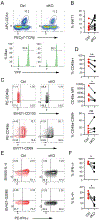
IFN-γ-producing invariant NKT (iNKT)1 cells are lipid-reactive innate-like lymphocytes that are resident in the thymus and peripheral tissues where they protect against pathogenic infection. The thymic functions of iNKT1 cells are not fully elucidated, but subsets of thymic iNKT cells modulate CD8 T cell, dendritic cell, B cell, and thymic epithelial cell numbers or function. In this study, we show that a subset of murine thymic iNKT1 cells required TGF-β-induced signals for their postselection development, to maintain hallmark TGF-β-induced genes, and for expression of the adhesion receptors CD49a and CD103. However, the residency-associated receptor CD69 was not TGF-β signaling-dependent. Recently described CD244+ c2 thymic iNKT1 cells, which produce IFN-γ without exogenous stimulation and have NK-like characteristics, reside in this TGF-β-responsive population. Liver and spleen iNKT1 cells do not share this TGF-β gene signature, but nonetheless TGF-β impacts liver iNKT1 cell phenotype and function. Our findings provide insight into the heterogeneity of mechanisms guiding iNKT1 cell development in different tissues and suggest a close association between a subset of iNKT1 cells and TGF-β-producing cells in the thymus that support their development.
Copyright © 2023 by The American Association of Immunologists, Inc.
Figures





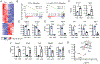
References
-
- Umeshappa CS, Sole P, Yamanouchi J, Mohapatra S, Surewaard BGJ, Garnica J, Singha S, Mondal D, Cortes-Vicente E, D'Mello C, Mason A, Kubes P, Serra P, Yang Y, and Santamaria P. 2022. Re-programming mouse liver-resident invariant natural killer T cells for suppressing hepatic and diabetogenic autoimmunity. Nat Commun 13: 3279. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

