The selected genes NR6A1, RSAD2-CMPK2, and COL3A1 contribute to body size variation in Meishan pigs through different patterns
- PMID: 37703114
- PMCID: PMC10548407
- DOI: 10.1093/jas/skad304
The selected genes NR6A1, RSAD2-CMPK2, and COL3A1 contribute to body size variation in Meishan pigs through different patterns
Abstract
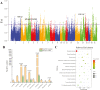
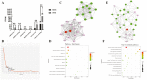
The high-fertility Meishan pig is currently categorized into medium sized (MMS) and small sized (SMS) based on body size. To identify causal genes responsible for the variation in body size within the two categories, we sequenced individuals representing the entire consanguinity of the existing Meishan pig. This enabled us to conduct genome selective signal analysis. Our findings revealed the genomes of MMS and SMS are stratified, with selective sweep regions formed by differential genomic intervals between the two categories enriched in multiple pig body size related quantitative trait loci (QTLs). Furthermore, the missense mutation c.575T > C of candidate causal gene NR6A1, accounting for the variation in lumbar vertebrae number in pigs, was positively selected in MMS only, leading to an increase in body length of MMS at 6 months of age. To precisely identify causal genes accounting for body size variation through multi-omics, we collected femoral cartilage and liver transcription data from MMS and SMS respectively, and re-sequencing data from pig breeds exhibiting varying body sizes. We found that two selected regions where the RSAD2-CMPK2 and COL3A1 genes are located, respectively, showed different haplotypes in pig breeds of varying body size, and was associated with body or carcass length in hybridized Suhuai pig. Additionally, the above three hub genes, were significantly greater expressed in SMS femoral cartilage and liver tissues compared to MMS. These three genes could strengthen the pathways related to bone resorption and metabolism in SMS, potentially hindering bone and skeletal development and resulting in a smaller body size in SMS. These findings provide valuable insights into the genetic mechanism of body size variation in Meishan pig population.
Keywords: COL3A1; NR6A1; RSAD2-CMPK2; Meishan pig; body size; bone resorption and digestion.
Plain language summary
The existing well-known Meishan pig population has been categorized into medium sized (MMS), and small sized (SMS) based on body size, which is a result of artificial selection. MMS is relatively large in all body size traits, but otherwise have highly similar appearance and performance traits. To effectively identify the candidate selected genes that contribute to the body size variation in Meishan pigs, this study collected individuals from all lineages of MMS and SMS for re-sequencing. Additionally, femoral cartilage and liver transcription data were collected from MMS and SMS, respectively, and re-sequencing data from pig breeds exhibiting varying body sizes were also analyzed. Through multi-omics analysis, it was discovered that the missense mutation c.575T > C in the candidate causal gene NR6A1 was positively selected in MMS only, leading to an increase in the body length of MMS at 6 months of age. Moreover, the selected genes RSAD2-CMPK2 and COL3A1 were found to be significantly greater expressed in SMS femoral cartilage and liver tissues compared with MMS. These genes could potentially strengthen bone resorption and metabolism-related pathways in SMS. These findings contribute to a better understanding of the genetic mechanisms underlying body size variation in Meishan pigs and Chinese indigenous pigs.
© The Author(s) 2023. Published by Oxford University Press on behalf of the American Society of Animal Science. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Genomic signatures reveal selection of characteristics within and between Meishan pig populations.Anim Genet. 2018 Apr;49(2):119-126. doi: 10.1111/age.12642. Epub 2018 Mar 6. Anim Genet. 2018. PMID: 29508928
-
Genetic evidence for the introgression of Western NR6A1 haplotype into Chinese Licha breed associated with increased vertebral number.Anim Genet. 2009 Apr;40(2):247-50. doi: 10.1111/j.1365-2052.2008.01820.x. Epub 2009 Feb 10. Anim Genet. 2009. PMID: 19220230
-
MC1R, KIT, IGF2, and NR6A1 as markers for genetic differentiation in Thai native, wild boars, and Duroc and Chinese Meishan pigs.Genet Mol Res. 2015 Oct 19;14(4):12723-32. doi: 10.4238/2015.October.19.16. Genet Mol Res. 2015. PMID: 26505423
-
Chromosome-Level Genome Assembly of the Meishan Pig and Insights into Its Domestication Mechanisms.Animals (Basel). 2025 Feb 19;15(4):603. doi: 10.3390/ani15040603. Animals (Basel). 2025. PMID: 40003085 Free PMC article.
-
A further survey of the quantitative trait loci affecting swine body size and carcass traits in five related pig populations.Anim Genet. 2021 Oct;52(5):621-632. doi: 10.1111/age.13112. Epub 2021 Jun 28. Anim Genet. 2021. PMID: 34182604
Cited by
-
Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China.Animals (Basel). 2024 Feb 11;14(4):594. doi: 10.3390/ani14040594. Animals (Basel). 2024. PMID: 38396562 Free PMC article. Review.
-
Muscle growth differences in Lijiang pigs revealed by ATAC-seq multi-omics.Front Vet Sci. 2024 Aug 26;11:1431248. doi: 10.3389/fvets.2024.1431248. eCollection 2024. Front Vet Sci. 2024. PMID: 39253524 Free PMC article.
-
Unexpected regulatory functions of cyprinid Viperin on inflammation and metabolism.BMC Genomics. 2024 Jun 29;25(1):650. doi: 10.1186/s12864-024-10566-x. BMC Genomics. 2024. PMID: 38951796 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

