Codependencies of mTORC1 signaling and endolysosomal actin structures
- PMID: 37703363
- PMCID: PMC10881074
- DOI: 10.1126/sciadv.add9084
Codependencies of mTORC1 signaling and endolysosomal actin structures
Abstract
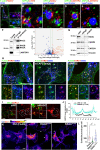
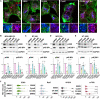
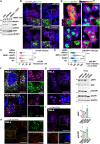
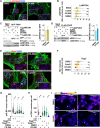
The mechanistic target of rapamycin complex 1 (mTORC1) is part of the amino acid sensing machinery that becomes activated on the endolysosomal surface in response to nutrient cues. Branched actin generated by WASH and Arp2/3 complexes defines endolysosomal microdomains. Here, we find mTORC1 components in close proximity to endolysosomal actin microdomains. We investigated for interactors of the mTORC1 lysosomal tether, RAGC, by proteomics and identified multiple actin filament capping proteins and their modulators. Perturbation of RAGC function affected the size of endolysosomal actin, consistent with a regulation of actin filament capping by RAGC. Reciprocally, the pharmacological inhibition of actin polymerization or alteration of endolysosomal actin obtained upon silencing of WASH or Arp2/3 complexes impaired mTORC1 activity. Mechanistically, we show that actin is required for proper association of RAGC and mTOR with endolysosomes. This study reveals an unprecedented interplay between actin and mTORC1 signaling on the endolysosomal system.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

