Chimeric hemagglutinin split vaccines elicit broadly cross-reactive antibodies and protection against group 2 influenza viruses in mice
- PMID: 37703367
- PMCID: PMC10499326
- DOI: 10.1126/sciadv.adi4753
Chimeric hemagglutinin split vaccines elicit broadly cross-reactive antibodies and protection against group 2 influenza viruses in mice
Abstract
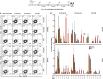
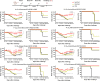
Seasonal influenza virus vaccines are effective when they are well matched to circulating strains. Because of antigenic drift/change in the immunodominant hemagglutinin (HA) head domain, annual vaccine reformulations are necessary to maintain a match with circulating strains. In addition, seasonal vaccines provide little to no protection against newly emerging pandemic strains. Sequential vaccination with chimeric HA (cHA) constructs has been proven to direct the immune response toward the immunosubdominant but more conserved HA stalk domain. In this study, we show that immunization with group 2 cHA split vaccines in combination with the CpG 1018 adjuvant elicits broadly cross-reactive antibodies against all group 2 HAs, as well as systemic and local antigen-specific T cell responses. Antibodies elicited after sequential vaccination are directed to conserved regions of the HA such as the stalk and the trimer interface and also to the N2 neuraminidase (NA). Immunized mice were fully protected from challenge with a broad panel of influenza A viruses.
Figures



Similar articles
-
Influenza H3 hemagglutinin vaccine with scrambled immunodominant epitopes elicits antibodies directed toward immunosubdominant head epitopes.mBio. 2023 Aug 31;14(4):e0062223. doi: 10.1128/mbio.00622-23. Epub 2023 Jul 19. mBio. 2023. PMID: 37466314 Free PMC article.
-
Neuraminidase-Inhibiting Antibody Titers Correlate with Protection from Heterologous Influenza Virus Strains of the Same Neuraminidase Subtype.J Virol. 2018 Aug 16;92(17):e01006-18. doi: 10.1128/JVI.01006-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29925654 Free PMC article.
-
Sequential Immunization With Live-Attenuated Chimeric Hemagglutinin-Based Vaccines Confers Heterosubtypic Immunity Against Influenza A Viruses in a Preclinical Ferret Model.Front Immunol. 2019 Apr 10;10:756. doi: 10.3389/fimmu.2019.00756. eCollection 2019. Front Immunol. 2019. PMID: 31105689 Free PMC article.
-
Towards a universal influenza vaccine: different approaches for one goal.Virol J. 2018 Jan 19;15(1):17. doi: 10.1186/s12985-017-0918-y. Virol J. 2018. PMID: 29370862 Free PMC article. Review.
-
Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease?Curr Opin Immunol. 2016 Oct;42:48-55. doi: 10.1016/j.coi.2016.05.012. Epub 2016 Jun 3. Curr Opin Immunol. 2016. PMID: 27268395 Free PMC article. Review.
Cited by
-
Sequential Immunizations with Influenza Neuraminidase Protein Followed by Peptide Nanoclusters Induce Heterologous Protection.Viruses. 2024 Jan 3;16(1):77. doi: 10.3390/v16010077. Viruses. 2024. PMID: 38257777 Free PMC article.
-
Multi-COBRA hemagglutinin formulated with cGAMP microparticles elicit protective immune responses against influenza viruses.bioRxiv [Preprint]. 2024 Feb 29:2024.02.27.582355. doi: 10.1101/2024.02.27.582355. bioRxiv. 2024. Update in: mSphere. 2024 Jul 30;9(7):e0016024. doi: 10.1128/msphere.00160-24. PMID: 38464191 Free PMC article. Updated. Preprint.
-
DNA and protein-generated chimeric molecules for delivery of influenza viral epitopes in mouse and humanized NSG transfer models.Hum Vaccin Immunother. 2024 Dec 31;20(1):2292381. doi: 10.1080/21645515.2023.2292381. Epub 2024 Jan 9. Hum Vaccin Immunother. 2024. PMID: 38193304 Free PMC article. Review.
-
Flublok Quadrivalent Vaccine Adjuvanted with R-DOTAP Elicits a Robust and Multifunctional CD4 T Cell Response That Is of Greater Magnitude and Functional Diversity Than Conventional Adjuvant Systems.Vaccines (Basel). 2024 Mar 7;12(3):281. doi: 10.3390/vaccines12030281. Vaccines (Basel). 2024. PMID: 38543915 Free PMC article.
-
The potential of nano- and microparticle-based influenza vaccines with enhanced shelf lives.Nanomedicine (Lond). 2024;19(26):2135-2138. doi: 10.1080/17435889.2024.2392484. Epub 2024 Oct 8. Nanomedicine (Lond). 2024. PMID: 39377099 No abstract available.
References
-
- E. A. Belongia, H. Q. McLean, Influenza Vaccine Effectiveness: Defining the H3N2 Problem. Clin. Infect. Dis. 69, 1817–1823 (2019). - PubMed
-
- G. N. Okoli, F. Racovitan, T. Abdulwahid, C. H. Righolt, S. M. Mahmud, Variable seasonal influenza vaccine effectiveness across geographical regions, age groups and levels of vaccine antigenic similarity with circulating virus strains: A systematic review and meta-analysis of the evidence from test-negative design studies after the 2009/10 influenza pandemic. Vaccine 39, 1225–1240 (2021). - PubMed
-
- Y. Suzuki, Positive selection for gains of N-linked glycosylation sites in hemagglutinin during evolution of H3N2 human influenza A virus. Genes Genet. Syst. 86, 287–294 (2011). - PubMed
-
- S. Rajaram, R. Wojcik, C. Moore, R. Ortiz de Lejarazu, S. de Lusignan, E. Montomoli, A. Rossi, A. Pérez-Rubio, A. Trilla, V. Baldo, R. Jandhyala, G. Kassianos, The impact of candidate influenza virus and egg-based manufacture on vaccine effectiveness: Literature review and expert consensus. Vaccine 38, 6047–6056 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

