Comparison of digital PCR platforms using the molecular marker
- PMID: 37704210
- PMCID: PMC10326530
- DOI: 10.5808/gi.23008
Comparison of digital PCR platforms using the molecular marker
Abstract
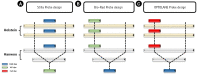
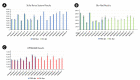
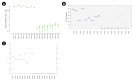
Assays of clinical diagnosis and species identification using molecular markers are performed according to a quantitative method in consideration of sensitivity, cost, speed, convenience, and specificity. However, typical polymerase chain reaction (PCR) assay is difficult to quantify and have various limitations. In addition, to perform quantitative analysis with the quantitative real-time PCR (qRT-PCR) equipment, a standard curve or normalization using reference genes is essential. Within the last a decade, previous studies have reported that the digital PCR (dPCR) assay, a third-generation PCR, can be applied in various fields by overcoming the shortcomings of typical PCR and qRT-PCR assays. We selected Stilla Naica System (Stilla Technologies), Droplet Digital PCR Technology (Bio-Rad), and Lab on an Array Digital Real-Time PCR analyzer system (OPTOLANE) for comparative analysis among the various droplet digital PCR platforms currently in use commercially. Our previous study discovered a molecular marker that can distinguish Hanwoo species (Korean native cattle) using Hanwoo-specific genomic structural variation. Here, we report the pros and cons of the operation of each dPCR platform from various perspectives using this species identification marker. In conclusion, we hope that this study will help researchers to select suitable dPCR platforms according to their purpose and resources.
Keywords: digital PCR platforms; molecular marker; point-of-care testing.
Conflict of interest statement
This study was supported by IL-YANG Pharmaceutical Co., Ltd.
Figures

References
-
- Hill WE. The polymerase chain reaction: applications for the detection of foodborne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. - PubMed
-
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. - PubMed
-
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. - PubMed
-
- Murphy J, Bustin SA. Reliability of real-time reverse-transcription PCR in clinical diagnostics: gold standard or substandard? Expert Rev Mol Diagn. 2009;9:187–197. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
