Early-Stage Breast Cancer Detection in Breast Milk
- PMID: 37704212
- PMCID: PMC10551665
- DOI: 10.1158/2159-8290.CD-22-1340
Early-Stage Breast Cancer Detection in Breast Milk
Abstract
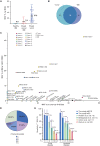
Breast cancer occurring during pregnancy (PrBC) and postpartum (PPBC) is usually diagnosed at more advanced stages compared with other breast cancer, worsening its prognosis. PPBC is particularly aggressive, with increased metastatic risk and mortality. Thus, effective screening methods to detect early PrBC and PPBC are needed. We report for the first time that cell-free tumor DNA (ctDNA) is present in breast milk (BM) collected from patients with breast cancer. Analysis of ctDNA from BM detects tumor variants in 87% of the cases by droplet digital PCR, while variants remain undetected in 92% of matched plasma samples. Retrospective next-generation sequencing analysis in BM ctDNA recapitulates tumor variants, with an overall clinical sensitivity of 71.4% and specificity of 100%. In two cases, ctDNA was detectable in BM collected 18 and 6 months prior to standard diagnosis. Our results open up the potential use of BM as a new source for liquid biopsy for PPBC detection.
Significance: For the first time, we show that BM obtained from patients with breast cancer carries ctDNA, surpassing plasma-based liquid biopsy for detection and molecular profiling of early-stage breast cancer, even prior to diagnosis by image. See related commentary by Cunningham and Turner, p. 2125. This article is featured in Selected Articles from This Issue, p. 2109.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
-
Liquid Biopsies in Breast Milk for the Early Detection of Breast Cancer.Cancer Discov. 2023 Oct 5;13(10):2125-2127. doi: 10.1158/2159-8290.CD-23-0836. Cancer Discov. 2023. PMID: 37794840
References
-
- Amant F, Loibl S, Neven P, Van Calsteren K. Malignancies in pregnancy 2 breast cancer in pregnancy. Lancet 2012;379:570–9. - PubMed
-
- Amant F, Von Minckwitz G, Han SN, Bontenbal M, Ring AE, Giermek J, et al. . Prognosis of women with primary breast cancer diagnosed during pregnancy: results from an international collaborative study. J Clin Oncol 2013;31:2532–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

