A foundation model for generalizable disease detection from retinal images
- PMID: 37704728
- PMCID: PMC10550819
- DOI: 10.1038/s41586-023-06555-x
A foundation model for generalizable disease detection from retinal images
Abstract
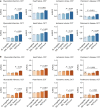
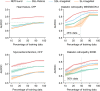
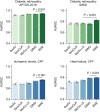
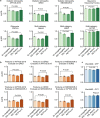
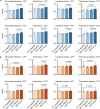
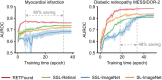
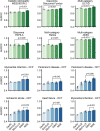
Medical artificial intelligence (AI) offers great potential for recognizing signs of health conditions in retinal images and expediting the diagnosis of eye diseases and systemic disorders1. However, the development of AI models requires substantial annotation and models are usually task-specific with limited generalizability to different clinical applications2. Here, we present RETFound, a foundation model for retinal images that learns generalizable representations from unlabelled retinal images and provides a basis for label-efficient model adaptation in several applications. Specifically, RETFound is trained on 1.6 million unlabelled retinal images by means of self-supervised learning and then adapted to disease detection tasks with explicit labels. We show that adapted RETFound consistently outperforms several comparison models in the diagnosis and prognosis of sight-threatening eye diseases, as well as incident prediction of complex systemic disorders such as heart failure and myocardial infarction with fewer labelled data. RETFound provides a generalizable solution to improve model performance and alleviate the annotation workload of experts to enable broad clinical AI applications from retinal imaging.
© 2023. The Author(s).
Conflict of interest statement
P.A.K. has acted as a consultant for DeepMind, Roche, Novartis, Apellis and BitFount, and is an equity owner in Big Picture Medical. He has received speaker fees from Heidelberg Engineering, Topcon, Allergan and Bayer.
Figures



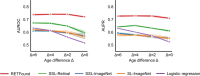
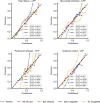
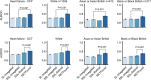
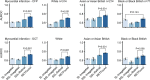
References
-
- Rajpurkar, P., Chen, E., Banerjee, O. & Topol, E. J. AI in health and medicine. Nat. Med.10.1038/s41591-021-01614-0 (2022). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

