SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes
- PMID: 37705046
- PMCID: PMC10500938
- DOI: 10.1186/s13287-023-03485-3
SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes
Abstract
Background: Cardiovascular complications significantly augment the overall COVID-19 mortality, largely due to the susceptibility of human cardiomyocytes (CMs) to SARS-CoV-2 virus. SARS-CoV-2 virus encodes 27 genes, whose specific impacts on CM health are not fully understood. This study elucidates the deleterious effects of SARS-CoV-2 genes Nsp6, M, and Nsp8 on human CMs.
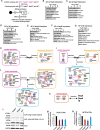
Methods: CMs were derived from human pluripotent stem cells (hPSCs), including human embryonic stem cells and induced pluripotent stem cells, using 2D and 3D differentiation methods. We overexpressed Nsp6, M, or Nsp8 in hPSCs and then applied whole mRNA-seq and mass spectrometry for multi-omics analysis. Co-immunoprecipitation mass spectrometry was utilized to map the protein interaction networks of Nsp6, M, and Nsp8 within host hiPSC-CMs.
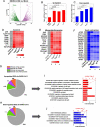
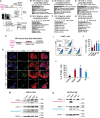
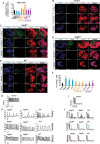
Results: Nsp6, Nsp8, and M globally perturb the transcriptome and proteome of hPSC-CMs. SARS-CoV-2 infection and the overexpression of Nsp6, Nsp8, or M coherently upregulated genes associated with apoptosis and immune/inflammation pathways, whereas downregulated genes linked to heart contraction and functions. Global interactome analysis revealed interactions between Nsp6, Nsp8, and M with ATPase subunits. Overexpression of Nsp6, Nsp8, or M significantly reduced cellular ATP levels, markedly increased apoptosis, and compromised Ca2+ handling in hPSC-CMs. Importantly, administration of FDA-approved drugs, ivermectin and meclizine, could restore ATP levels, thereby mitigating apoptosis and dysfunction in hPSC-CMs overexpressing Nsp6, Nsp8, or M.
Conclusion: Overall, our findings uncover the extensive damaging effects of Nsp6, Nsp8, and M on hPSC-CMs, underlining the crucial role of ATP homeostasis in CM death and functional abnormalities induced by these SARS-CoV-2 genes, and reveal the potential therapeutic strategies to alleviate these detrimental effects with FDA-approved drugs.
Keywords: Apoptosis; COVID-19; Cardiac dysfunction; Cardiomyocyte; Human pluripotent stem cells; SARS-CoV-2.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide analyses reveal the detrimental impacts of SARS-CoV-2 viral gene Orf9c on human pluripotent stem cell-derived cardiomyocytes.Stem Cell Reports. 2022 Mar 8;17(3):522-537. doi: 10.1016/j.stemcr.2022.01.014. Epub 2022 Feb 17. Stem Cell Reports. 2022. PMID: 35180394 Free PMC article.
-
Decoding HiPSC-CM's Response to SARS-CoV-2: mapping the molecular landscape of cardiac injury.BMC Genomics. 2024 Mar 12;25(1):271. doi: 10.1186/s12864-024-10194-5. BMC Genomics. 2024. PMID: 38475718 Free PMC article.
-
SARS-CoV-2 Infects Human Pluripotent Stem Cell-Derived Cardiomyocytes, Impairing Electrical and Mechanical Function.Stem Cell Reports. 2021 Mar 9;16(3):478-492. doi: 10.1016/j.stemcr.2021.02.008. Epub 2021 Feb 13. Stem Cell Reports. 2021. PMID: 33657418 Free PMC article.
-
Modeling Cardiac SARS-CoV-2 Infection with Human Pluripotent Stem Cells.Curr Cardiol Rep. 2022 Dec;24(12):2121-2129. doi: 10.1007/s11886-022-01813-2. Epub 2022 Oct 22. Curr Cardiol Rep. 2022. PMID: 36272051 Free PMC article. Review.
-
Pluripotent stem cell-based cardiac regenerative therapy for heart failure.J Mol Cell Cardiol. 2024 Feb;187:90-100. doi: 10.1016/j.yjmcc.2023.12.001. Epub 2024 Feb 6. J Mol Cell Cardiol. 2024. PMID: 38331557 Review.
Cited by
-
An In-Depth Insight into Clinical, Cellular and Molecular Factors in COVID19-Associated Cardiovascular Ailments for Identifying Novel Disease Biomarkers, Drug Targets and Clinical Management Strategies.Arch Microbiol Immunol. 2024;8(3):290-308. doi: 10.26502/ami.936500177. Epub 2024 Jul 19. Arch Microbiol Immunol. 2024. PMID: 39703820 Free PMC article.
-
NSP6 regulates calcium overload-induced autophagic cell death and is regulated by KLHL22-mediated ubiquitination.J Adv Res. 2025 Aug;74:303-318. doi: 10.1016/j.jare.2025.05.031. Epub 2025 May 13. J Adv Res. 2025. PMID: 40373961 Free PMC article.
-
Impact of SARS-CoV-2 Wuhan and Omicron Variant Proteins on Type I Interferon Response.Viruses. 2025 Apr 15;17(4):569. doi: 10.3390/v17040569. Viruses. 2025. PMID: 40285011 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

