Increased expression of BRD4 isoforms long (BRD4-L) and short (BRD4-S) promotes chemotherapy resistance in high-grade serous ovarian carcinoma
- PMID: 37705995
- PMCID: PMC10496930
- DOI: 10.18632/genesandcancer.233
Increased expression of BRD4 isoforms long (BRD4-L) and short (BRD4-S) promotes chemotherapy resistance in high-grade serous ovarian carcinoma
Abstract
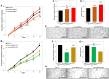
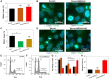
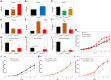
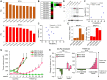
Chemoresistance in ovarian carcinoma is a puzzling issue that urges understanding of strategies used by cancer cells to survive DNA damage and to escape cell death. Expanding efforts to understand mechanisms driving chemoresistance and to develop alternative therapies targeting chemoresistant tumors are critical. Amplification of BRD4 is frequently associated with chemoresistant ovarian carcinoma, but little is known about the biological effects of the overexpression of BRD4 isoforms in this malignancy. Here, we described the consequences of BRD4-L and BRD4-S overexpression in ovarian carcinoma shedding a light on a complex regulation of BRD4 isoforms. We demonstrated that the BRD4-L transcript expression is required to generate both isoforms, BRD4-L and BRD4-S. We showed that the BRD4-S mRNA expression positively correlated with BRD4-S protein levels, while BRD4-L isoform showed negative correlation between mRNA and protein levels. Moreover, we demonstrated that an overexpression of BRD4 isoforms is associated with chemoresistance in ovarian cancer.
Keywords: BRD4 amplification; BRD4 long; BRD4 short; chemoresistance; high-grade serous ovarian carcinoma.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that there are no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

