Transcriptional changes of the aging lung
- PMID: 37706427
- PMCID: PMC10577555
- DOI: 10.1111/acel.13969
Transcriptional changes of the aging lung
Abstract
Aging is a natural process associated with declined organ function and higher susceptibility to developing chronic diseases. A systemic single-cell type-based study provides a unique opportunity to understand the mechanisms behind age-related pathologies. Here, we use single-cell gene expression analysis comparing healthy young and aged human lungs from nonsmoker donors to investigate age-related transcriptional changes. Our data suggest that aging has a heterogenous effect on lung cells, as some populations are more transcriptionally dynamic while others remain stable in aged individuals. We found that monocytes and alveolar macrophages were the most transcriptionally affected populations. These changes were related to inflammation and regulation of the immune response. Additionally, we calculated the LungAge score, which reveals the diversity of lung cell types during aging. Changes in DNA damage repair, fatty acid metabolism, and inflammation are essential for age prediction. Finally, we quantified the senescence score in aged lungs and found that the more biased cells toward senescence are immune and progenitor cells. Our study provides a comprehensive and systemic analysis of the molecular signatures of lung aging. Our LungAge signature can be used to predict molecular signatures of physiological aging and to detect common signatures of age-related lung diseases.
Keywords: aging; inflammation; lung; senescence; single-cell RNA-seq.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors state no conflict of interest.
Figures
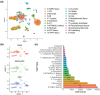



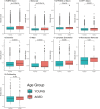

References
-
- Adams, T. S. , Schupp, J. C. , Poli, S. , Ayaub, E. A. , Neumark, N. , Ahangari, F. , Chu, S. G. , Raby, B. A. , DeIuliis, G. , Januszyk, M. , Duan, Q. , Arnett, H. A. , Siddiqui, A. , Washko, G. R. , Homer, R. , Yan, X. , Rosas, I. O. , & Kaminski, N. (2020). Single‐cell RNA‐seq reveals ectopic and aberrant lung‐resident cell populations in idiopathic pulmonary fibrosis. Science Advances, 6(28), eaba1983. - PMC - PubMed
-
- Agusti, A. , & Faner, R. (2019). Lung function trajectories in health and disease. The Lancet Respiratory Medicine, 7(4), 358–364. - PubMed
-
- Almanan, M. , Raynor, J. , Ogunsulire, I. , Malyshkina, A. , Mukherjee, S. , Hummel, S. A. , Ingram, J. T. , Saini, A. , Xie, M. M. , Alenghat, T. , Way, S. S. , Deepe, G. S., Jr. , Divanovic, S. , Singh, H. , Miraldi, E. , Zajac, A. J. , Dent, A. L. , Holscher, C. , Chougnet, C. , & Hildeman, D. A. (2020). IL‐10‐producing Tfh cells accumulate with age and link inflammation with age‐related immune suppression. Science Advances, 6(31), eabb0806. - PMC - PubMed
-
- DePianto, D. J. , Heiden, J. A. V. , Morshead, K. B. , Sun, K. H. , Modrusan, Z. , Teng, G. , Wolters, P. J. , & Arron, J. R. (2021). Molecular mapping of interstitial lung disease reveals a phenotypically distinct senescent basal epithelial cell population. JCI Insight, 6(8), e143626. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

