A cyclic pyrrole-imidazole polyamide reduces pathogenic RNA in CAG/CTG triplet repeat neurological disease models
- PMID: 37707954
- PMCID: PMC10645379
- DOI: 10.1172/JCI164792
A cyclic pyrrole-imidazole polyamide reduces pathogenic RNA in CAG/CTG triplet repeat neurological disease models
Abstract
Expansion of CAG and CTG (CWG) triplet repeats causes several inherited neurological diseases. The CWG repeat diseases are thought to involve complex pathogenic mechanisms through expanded CWG repeat-derived RNAs in a noncoding region and polypeptides in a coding region, respectively. However, an effective therapeutic approach has not been established for the CWG repeat diseases. Here, we show that a CWG repeat DNA-targeting compound, cyclic pyrrole-imidazole polyamide (CWG-cPIP), suppressed the pathogenesis of coding and noncoding CWG repeat diseases. CWG-cPIP bound to the hairpin form of mismatched CWG DNA, interfering with transcription elongation by RNA polymerase through a preferential activity toward repeat-expanded DNA. We found that CWG-cPIP selectively inhibited pathogenic mRNA transcripts from expanded CWG repeats, reducing CUG RNA foci and polyglutamine accumulation in cells from patients with myotonic dystrophy type 1 (DM1) and Huntington's disease (HD). Treatment with CWG-cPIP ameliorated behavioral deficits in adeno-associated virus-mediated CWG repeat-expressing mice and in a genetic mouse model of HD, without cytotoxicity or off-target effects. Together, we present a candidate compound that targets expanded CWG repeat DNA independently of its genomic location and reduces both pathogenic RNA and protein levels. CWG-cPIP may be used for the treatment of CWG repeat diseases and improvement of clinical outcomes.
Keywords: Genetic diseases; Neuroscience; Noncoding RNAs; Therapeutics; Transcription.
Figures

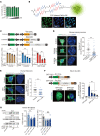

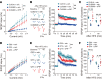
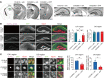
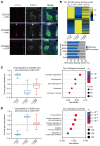
Comment in
-
Targeting expanded trinucleotide repeats.Nat Rev Drug Discov. 2023 Nov;22(11):873. doi: 10.1038/d41573-023-00160-3. Nat Rev Drug Discov. 2023. PMID: 37798465 No abstract available.
Similar articles
-
Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells.Hum Mol Genet. 2013 Dec 20;22(25):5276-87. doi: 10.1093/hmg/ddt386. Epub 2013 Aug 9. Hum Mol Genet. 2013. PMID: 23933738 Free PMC article.
-
Strong and Specific Recognition of CAG/CTG Repeat DNA (5'-dWGCWGCW-3') by a Cyclic Pyrrole-Imidazole Polyamide.Chembiochem. 2022 Jan 19;23(2):e202100533. doi: 10.1002/cbic.202100533. Epub 2021 Nov 18. Chembiochem. 2022. PMID: 34796607 Free PMC article.
-
Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy.PLoS Genet. 2011 Mar;7(3):e1001340. doi: 10.1371/journal.pgen.1001340. Epub 2011 Mar 17. PLoS Genet. 2011. PMID: 21437269 Free PMC article.
-
CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders.RNA Biol. 2011 Jul-Aug;8(4):565-71. doi: 10.4161/rna.8.4.15397. Epub 2011 Jul 1. RNA Biol. 2011. PMID: 21593608 Free PMC article. Review.
-
CRISPR/Cas Applications in Myotonic Dystrophy: Expanding Opportunities.Int J Mol Sci. 2019 Jul 27;20(15):3689. doi: 10.3390/ijms20153689. Int J Mol Sci. 2019. PMID: 31357652 Free PMC article. Review.
Cited by
-
Therapeutic approaches targeting aging and cellular senescence in Huntington's disease.CNS Neurosci Ther. 2024 Oct;30(10):e70053. doi: 10.1111/cns.70053. CNS Neurosci Ther. 2024. PMID: 39428700 Free PMC article. Review.
-
Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.JACS Au. 2024 May 15;4(5):1801-1810. doi: 10.1021/jacsau.3c00830. eCollection 2024 May 27. JACS Au. 2024. PMID: 38818057 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

