DNA-methylation signature accurately differentiates pancreatic cancer from chronic pancreatitis in tissue and plasma
- PMID: 37709492
- PMCID: PMC10715533
- DOI: 10.1136/gutjnl-2023-330155
DNA-methylation signature accurately differentiates pancreatic cancer from chronic pancreatitis in tissue and plasma
Abstract
Objective: Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy. Differentiation from chronic pancreatitis (CP) is currently inaccurate in about one-third of cases. Misdiagnoses in both directions, however, have severe consequences for patients. We set out to identify molecular markers for a clear distinction between PDAC and CP.
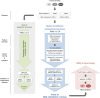
Design: Genome-wide variations of DNA-methylation, messenger RNA and microRNA level as well as combinations thereof were analysed in 345 tissue samples for marker identification. To improve diagnostic performance, we established a random-forest machine-learning approach. Results were validated on another 48 samples and further corroborated in 16 liquid biopsy samples.
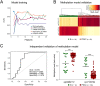
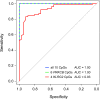
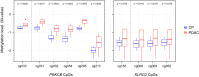
Results: Machine-learning succeeded in defining markers to differentiate between patients with PDAC and CP, while low-dimensional embedding and cluster analysis failed to do so. DNA-methylation yielded the best diagnostic accuracy by far, dwarfing the importance of transcript levels. Identified changes were confirmed with data taken from public repositories and validated in independent sample sets. A signature of six DNA-methylation sites in a CpG-island of the protein kinase C beta type gene achieved a validated diagnostic accuracy of 100% in tissue and in circulating free DNA isolated from patient plasma.
Conclusion: The success of machine-learning to identify an effective marker signature documents the power of this approach. The high diagnostic accuracy of discriminating PDAC from CP could have tremendous consequences for treatment success, once the result from still a limited number of liquid biopsy samples would be confirmed in a larger cohort of patients with suspected pancreatic cancer.
Keywords: chronic pancreatitis; pancreatic cancer.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
