Stromal heterogeneity may explain increased incidence of metaplastic breast cancer in women of African descent
- PMID: 37709737
- PMCID: PMC10502140
- DOI: 10.1038/s41467-023-41473-6
Stromal heterogeneity may explain increased incidence of metaplastic breast cancer in women of African descent
Abstract
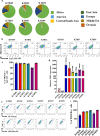
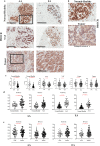
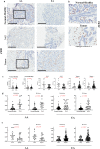
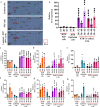
The biologic basis of genetic ancestry-dependent variability in disease incidence and outcome is just beginning to be explored. We recently reported enrichment of a population of ZEB1-expressing cells located adjacent to ductal epithelial cells in normal breasts of women of African ancestry compared to those of European ancestry. In this study, we demonstrate that these cells have properties of fibroadipogenic/mesenchymal stromal cells that express PROCR and PDGFRα and transdifferentiate into adipogenic and osteogenic lineages. PROCR + /ZEB1 + /PDGFRα+ (PZP) cells are enriched in normal breast tissues of women of African compared to European ancestry. PZP: epithelial cell communication results in luminal epithelial cells acquiring basal cell characteristics and IL-6-dependent increase in STAT3 phosphorylation. Furthermore, level of phospho-STAT3 is higher in normal and cancerous breast tissues of women of African ancestry. PZP cells transformed with HRasG12V ± SV40-T/t antigens generate metaplastic carcinoma suggesting that these cells are one of the cells-of-origin of metaplastic breast cancers.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

