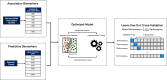
Genetic risk assessment based on association and prediction studies
- PMID: 37709797
- PMCID: PMC10502006
- DOI: 10.1038/s41598-023-41862-3
Genetic risk assessment based on association and prediction studies
Abstract
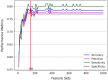
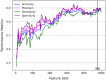
The genetic basis of phenotypic emergence provides valuable information for assessing individual risk. While association studies have been pivotal in identifying genetic risk factors within a population, complementing it with insights derived from predictions studies that assess individual-level risk offers a more comprehensive approach to understanding phenotypic expression. In this study, we established personalized risk assessment models using single-nucleotide polymorphism (SNP) data from 200 Korean patients, of which 100 experienced hepatitis B surface antigen (HBsAg) seroclearance and 100 patients demonstrated high levels of HBsAg. The risk assessment models determined the predictive power of the following: (1) genome-wide association study (GWAS)-identified candidate biomarkers considered significant in a reference study and (2) machine learning (ML)-identified candidate biomarkers with the highest feature importance scores obtained by using random forest (RF). While utilizing all features yielded 64% model accuracy, using relevant biomarkers achieved higher model accuracies: 82% for 52 GWAS-identified candidate biomarkers, 71% for three GWAS-identified biomarkers, and 80% for 150 ML-identified candidate biomarkers. Findings highlight that the joint contributions of relevant biomarkers significantly influence phenotypic emergence. On the other hand, combining ML-identified candidate biomarkers into the pool of GWAS-identified candidate biomarkers resulted in the improved predictive accuracy of 90%, demonstrating the capability of ML as an auxiliary analysis to GWAS. Furthermore, some of the ML-identified candidate biomarkers were found to be linked with hepatocellular carcinoma (HCC), reinforcing previous claims that HCC can still occur despite the absence of HBsAg.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A machine learning-based SNP-set analysis approach for identifying disease-associated susceptibility loci.Sci Rep. 2022 Sep 22;12(1):15817. doi: 10.1038/s41598-022-19708-1. Sci Rep. 2022. PMID: 36138111 Free PMC article.
-
A risk prediction model for hepatocellular carcinoma after hepatitis B surface antigen seroclearance.J Hepatol. 2022 Sep;77(3):632-641. doi: 10.1016/j.jhep.2022.03.032. Epub 2022 Apr 7. J Hepatol. 2022. PMID: 35398462
-
Genome-Wide Association Study Identifies TLL1 Variant Associated With Development of Hepatocellular Carcinoma After Eradication of Hepatitis C Virus Infection.Gastroenterology. 2017 May;152(6):1383-1394. doi: 10.1053/j.gastro.2017.01.041. Epub 2017 Feb 3. Gastroenterology. 2017. PMID: 28163062
-
What can we learn from hepatitis B virus clinical cohorts?Liver Int. 2015 Jan;35 Suppl 1:91-9. doi: 10.1111/liv.12716. Liver Int. 2015. PMID: 25529093 Review.
-
Association Between Seroclearance of Hepatitis B Surface Antigen and Long-term Clinical Outcomes of Patients With Chronic Hepatitis B Virus Infection: Systematic Review and Meta-analysis.Clin Gastroenterol Hepatol. 2021 Mar;19(3):463-472. doi: 10.1016/j.cgh.2020.05.041. Epub 2020 May 27. Clin Gastroenterol Hepatol. 2021. PMID: 32473348
Cited by
-
Temporal Trends in the Completeness of Epidemiological Variables in a Hospital-Based Cancer Registry of a Pediatric Oncology Center in Brazil.Int J Environ Res Public Health. 2024 Feb 9;21(2):200. doi: 10.3390/ijerph21020200. Int J Environ Res Public Health. 2024. PMID: 38397690 Free PMC article.
-
The classification method of donkey breeds based on SNPs data and machine learning.Front Genet. 2025 Apr 9;16:1496246. doi: 10.3389/fgene.2025.1496246. eCollection 2025. Front Genet. 2025. PMID: 40270545 Free PMC article.
-
Diagnostic performance of AI-based models versus physicians among patients with hepatocellular carcinoma: a systematic review and meta-analysis.Front Artif Intell. 2024 Aug 19;7:1398205. doi: 10.3389/frai.2024.1398205. eCollection 2024. Front Artif Intell. 2024. PMID: 39224209 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical