Pipeline validation for the identification of antimicrobial-resistant genes in carbapenem-resistant Klebsiella pneumoniae
- PMID: 37709838
- PMCID: PMC10502106
- DOI: 10.1038/s41598-023-42154-6
Pipeline validation for the identification of antimicrobial-resistant genes in carbapenem-resistant Klebsiella pneumoniae
Abstract
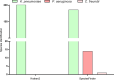
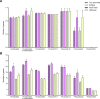
Antimicrobial-resistant Klebsiella pneumoniae is a global threat to healthcare and an important cause of nosocomial infections. Antimicrobial resistance causes prolonged treatment periods, high mortality rates, and economic impacts. Whole Genome Sequencing (WGS) has been used in laboratory diagnosis, but there is limited evidence about pipeline validation to parse generated data. Thus, the present study aimed to validate a bioinformatics pipeline for the identification of antimicrobial resistance genes from carbapenem-resistant K. pneumoniae WGS. Sequences were obtained from a publicly available database, trimmed, de novo assembled, mapped to the K. pneumoniae reference genome, and annotated. Contigs were submitted to different tools for bacterial (Kraken2 and SpeciesFinder) and antimicrobial resistance gene identification (ResFinder and ABRicate). We analyzed 201 K. pneumoniae genomes. In the bacterial identification by Kraken2, all samples were correctly identified, and in SpeciesFinder, 92.54% were correctly identified as K. pneumoniae, 6.96% erroneously as Pseudomonas aeruginosa, and 0.5% erroneously as Citrobacter freundii. ResFinder found a greater number of antimicrobial resistance genes than ABRicate; however, many were identified more than once in the same sample. All tools presented 100% repeatability and reproducibility and > 75% performance in other metrics. Kraken2 was more assertive in recognizing bacterial species, and SpeciesFinder may need improvements.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

