Allele mining of eukaryotic translation initiation factor genes in Prunus for the identification of new sources of resistance to sharka
- PMID: 37709842
- PMCID: PMC10502034
- DOI: 10.1038/s41598-023-42215-w
Allele mining of eukaryotic translation initiation factor genes in Prunus for the identification of new sources of resistance to sharka
Abstract
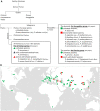
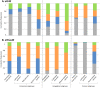
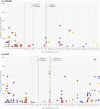
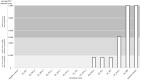
Members of the eukaryotic translation initiation complex are co-opted in viral infection, leading to susceptibility in many crop species, including stone fruit trees (Prunus spp.). Therefore, modification of one of those eukaryotic translation initiation factors or changes in their gene expression may result in resistance. We searched the crop and wild Prunus germplasm from the Armeniaca and Amygdalus taxonomic sections for allelic variants in the eIF4E and eIFiso4E genes, to identify alleles potentially linked to resistance to Plum pox virus (PPV). Over one thousand stone fruit accessions (1397) were screened for variation in eIF4E and eIFiso4E transcript sequences which are in single copy within the diploid Prunus genome. We identified new alleles for both genes differing from haplotypes associated with PPV susceptible accessions. Overall, analyses showed that eIFiso4E is genetically more constrained since it displayed less polymorphism than eIF4E. We also demonstrated more variations at both loci in the related wild species than in crop species. As the eIFiso4E translation initiation factor was identified as indispensable for PPV infection, a selection of ten different eIFiso4E haplotypes along 13 accessions were tested by infection with PPV and eight of them displayed a range of reduced susceptibility to resistance, indicating new potential sources of resistance to sharka.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Gómez P, Rodríguez-Hernández AM, Moury B, Aranda MA. Genetic resistance for the sustainable control of plant virus diseases: Breeding, mechanisms and durability. Eur. J. Plant Pathol. 2009;125:1–22. doi: 10.1007/s10658-009-9468-5. - DOI
-
- Fraser RSS. The genetics of resistance to plant viruses. Annu. Rev. Phytopathol. 1990;28:179–200. doi: 10.1146/annurev.py.28.090190.001143. - DOI
-
- Le Gall O, Aranda MA, Caranta C. Plant resistance to viruses mediated by translation initiation factors. In: Caranta C, Aranda MA, Tepfer M, Lopez-Moya JJ, editors. Recent Advances in Plant Virology. Caister Academic Press; 2011. pp. 177–194.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

