Genome-wide association meta-analysis identifies 17 loci associated with nonalcoholic fatty liver disease
- PMID: 37709864
- PMCID: PMC10918428
- DOI: 10.1038/s41588-023-01497-6
Genome-wide association meta-analysis identifies 17 loci associated with nonalcoholic fatty liver disease
Abstract
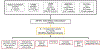
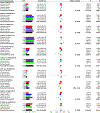
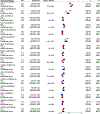
Nonalcoholic fatty liver disease (NAFLD) is common and partially heritable and has no effective treatments. We carried out a genome-wide association study (GWAS) meta-analysis of imaging (n = 66,814) and diagnostic code (3,584 cases versus 621,081 controls) measured NAFLD across diverse ancestries. We identified NAFLD-associated variants at torsin family 1 member B (TOR1B), fat mass and obesity associated (FTO), cordon-bleu WH2 repeat protein like 1 (COBLL1)/growth factor receptor-bound protein 14 (GRB14), insulin receptor (INSR), sterol regulatory element-binding transcription factor 1 (SREBF1) and patatin-like phospholipase domain-containing protein 2 (PNPLA2), as well as validated NAFLD-associated variants at patatin-like phospholipase domain-containing protein 3 (PNPLA3), transmembrane 6 superfamily 2 (TM6SF2), apolipoprotein E (APOE), glucokinase regulator (GCKR), tribbles homolog 1 (TRIB1), glycerol-3-phosphate acyltransferase (GPAM), mitochondrial amidoxime-reducing component 1 (MARC1), microsomal triglyceride transfer protein large subunit (MTTP), alcohol dehydrogenase 1B (ADH1B), transmembrane channel like 4 (TMC4)/membrane-bound O-acyltransferase domain containing 7 (MBOAT7) and receptor-type tyrosine-protein phosphatase δ (PTPRD). Implicated genes highlight mitochondrial, cholesterol and de novo lipogenesis as causally contributing to NAFLD predisposition. Phenome-wide association study (PheWAS) analyses suggest at least seven subtypes of NAFLD. Individuals in the top 10% and 1% of genetic risk have a 2.5-fold to 6-fold increased risk of NAFLD, cirrhosis and hepatocellular carcinoma. These genetic variants identify subtypes of NAFLD, improve estimates of disease risk and can guide the development of targeted therapeutics.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The Regents of the University of Michigan and E.K.S. have a pending patent on the use of systems and methods for analysis of samples associated with NAFLD and related conditions. V.L.C. received grant funding from KOWA and AstraZeneca. J.J.C. and Vanderbilt University Medical Center receive research funding from NIH, IBM Watson Health, GE Healthcare and Theratechnologies. G.R.W. is a cofounder and equity shareholder in Quantitative Imaging Solutions, a company that provides consulting services for image and data analytics. G.R.W.’s spouse works for Biogen. Grants or contracts from NIH, Department of Defense (DoD) and Boehringer Ingelheim made payments to G.R.W.’s institution. G.R.W. received consulting fees from Pulmonx, Vertex, Janssen Pharmaceuticals, Pieris Therapeutics and Intellia Therapeutics. G.R.W. also received payments from Pulmonx for participation on a Data Safety Monitoring Board or Advisory Board. The remaining authors declare no competing interests.
Figures
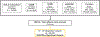
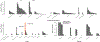


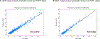


References
-
- Lauridsen BK et al. Liver fat content, non-alcoholic fatty liver disease, and ischaemic heart disease: Mendelian randomization and meta-analysis of 279 013 individuals. Eur. Heart J. 39, 385–393 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK131787/DK/NIDDK NIH HHS/United States
- R01 HL087660/HL/NHLBI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- HHSN271201200022C/DA/NIDA NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
- R01 HL122464/HL/NHLBI NIH HHS/United States
- R01 DK085175/DK/NIDDK NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- R01 HL071739/HL/NHLBI NIH HHS/United States
- HHSN268201800012C/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- R01 HL061210/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 DK106621/DK/NIDDK NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- HHSN268201800014I/HB/NHLBI NIH HHS/United States
- U01 HL137181/HL/NHLBI NIH HHS/United States
- HHSN268201800014C/HL/NHLBI NIH HHS/United States
- R01 HL060944/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- N01AG12100/AG/NIA NIH HHS/United States
- U01 HL084756/HL/NHLBI NIH HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201800013I/MD/NIMHD NIH HHS/United States
- RC1 HL100245/HL/NHLBI NIH HHS/United States
- R01 HL061019/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201800012I/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- U01 HL072515/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- HHSN268201800011C/HL/NHLBI NIH HHS/United States
- R01 DK107904/DK/NIDDK NIH HHS/United States
- R01 DK089256/DK/NIDDK NIH HHS/United States
- R01 DK118062/DK/NIDDK NIH HHS/United States
- R01 HL060919/HL/NHLBI NIH HHS/United States
- R01 HL085571/HL/NHLBI NIH HHS/United States
- HHSN268201800015I/HB/NHLBI NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- HHSN268201800010I/HB/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- R01 DK128871/DK/NIDDK NIH HHS/United States
- R01 HL117078/HL/NHLBI NIH HHS/United States
- R01 HL060894/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- HHSN268201800011I/HB/NHLBI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- R01 HL087700/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

