Targeted genome editing with a DNA-dependent DNA polymerase and exogenous DNA-containing templates
- PMID: 37709915
- PMCID: PMC12054351
- DOI: 10.1038/s41587-023-01947-w
Targeted genome editing with a DNA-dependent DNA polymerase and exogenous DNA-containing templates
Abstract
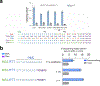
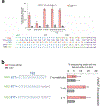
Reverse transcriptases, used in prime editing systems, exhibit lower fidelity, processivity and dNTP affinity than many DNA-dependent DNA polymerases. We report that a DNA-dependent DNA polymerase (phi29), untethered from Cas9, enables editing from a synthetic, end-stabilized DNA-containing template at up to 60% efficiency in human cells. Compared to prime editing, DNA polymerase editing avoids autoinhibitory intramolecular base pairing of the template, facilitates template synthesis and supports larger insertions (>100 nucleotides).
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
E.J.S. is a co-founder and Scientific Advisory Board member of Intellia Therapeutics and a Scientific Advisory Board member at Tessera Therapeutics. The University of Massachusetts Chan Medical School has filed patent applications related to this work. The authors declare no other competing interests.
Figures


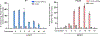


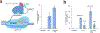




References
-
- Lin Q et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021). - PubMed
-
- Zhuang Y et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 18, 29–37 (2022). - PubMed
-
- Wang J et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 19, 331–340 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

