Human GLP1R variants affecting GLP1R cell surface expression are associated with impaired glucose control and increased adiposity
- PMID: 37709961
- PMCID: PMC11610247
- DOI: 10.1038/s42255-023-00889-6
Human GLP1R variants affecting GLP1R cell surface expression are associated with impaired glucose control and increased adiposity
Abstract
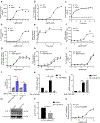
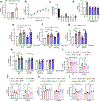
The glucagon-like peptide 1 receptor (GLP1R) is a major drug target with several agonists being prescribed in individuals with type 2 diabetes and obesity1,2. The impact of genetic variability of GLP1R on receptor function and its association with metabolic traits are unclear with conflicting reports. Here, we show an unexpected diversity of phenotypes ranging from defective cell surface expression to complete or pathway-specific gain of function (GoF) and loss of function (LoF), after performing a functional profiling of 60 GLP1R variants across four signalling pathways. The defective insulin secretion of GLP1R LoF variants is rescued by allosteric GLP1R ligands or high concentrations of exendin-4/semaglutide in INS-1 823/3 cells. Genetic association studies in 200,000 participants from the UK Biobank show that impaired GLP1R cell surface expression contributes to poor glucose control and increased adiposity with increased glycated haemoglobin A1c and body mass index. This study defines impaired GLP1R cell surface expression as a risk factor for traits associated with type 2 diabetes and obesity and provides potential treatment options for GLP1R LoF variant carriers.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interes
Figures



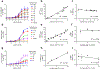















References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

