Overestimated prediction using polygenic prediction derived from summary statistics
- PMID: 37710206
- PMCID: PMC10500750
- DOI: 10.1186/s12863-023-01151-4
Overestimated prediction using polygenic prediction derived from summary statistics
Abstract
Background: When polygenic risk score (PRS) is derived from summary statistics, independence between discovery and test sets cannot be monitored. We compared two types of PRS studies derived from raw genetic data (denoted as rPRS) and the summary statistics for IGAP (sPRS).
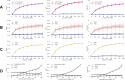
Results: Two variables with the high heritability in UK Biobank, hypertension, and height, are used to derive an exemplary scale effect of PRS. sPRS without APOE is derived from International Genomics of Alzheimer's Project (IGAP), which records ΔAUC and ΔR2 of 0.051 ± 0.013 and 0.063 ± 0.015 for Alzheimer's Disease Sequencing Project (ADSP) and 0.060 and 0.086 for Accelerating Medicine Partnership - Alzheimer's Disease (AMP-AD). On UK Biobank, rPRS performances for hypertension assuming a similar size of discovery and test sets are 0.0036 ± 0.0027 (ΔAUC) and 0.0032 ± 0.0028 (ΔR2). For height, ΔR2 is 0.029 ± 0.0037.
Conclusion: Considering the high heritability of hypertension and height of UK Biobank and sample size of UK Biobank, sPRS results from AD databases are inflated. Independence between discovery and test sets is a well-known basic requirement for PRS studies. However, a lot of PRS studies cannot follow such requirements because of impossible direct comparisons when using summary statistics. Thus, for sPRS, potential duplications should be carefully considered within the same ethnic group.
Keywords: Alzheimer’s disease; Complex genetic disease; Overestimation bias; Polygenic risk score.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
