Characterization of norbelladine synthase and noroxomaritidine/norcraugsodine reductase reveals a novel catalytic route for the biosynthesis of Amaryllidaceae alkaloids including the Alzheimer's drug galanthamine
- PMID: 37711303
- PMCID: PMC10499049
- DOI: 10.3389/fpls.2023.1231809
Characterization of norbelladine synthase and noroxomaritidine/norcraugsodine reductase reveals a novel catalytic route for the biosynthesis of Amaryllidaceae alkaloids including the Alzheimer's drug galanthamine
Abstract
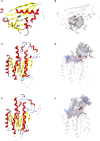
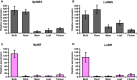
Amaryllidaceae alkaloids (AAs) are a large group of plant specialized metabolites with diverse pharmacological properties. Norbelladine is the entry compound in AAs biosynthesis and is produced from the condensation of tyramine and 3,4-dihydroxybenzaldehyde (3,4-DHBA). There are two reported enzymes capable of catalyzing this reaction in-vitro, both with low yield. The first one, norbelladine synthase (NBS), was shown to condense tyramine and 3,4-DHBA, while noroxomaritidine/norcraugsodine reductase (NR), catalyzes a reduction reaction to produce norbelladine. To clarify the mechanisms involved in this controversial step, both NBS and NR homologs were identified from the transcriptome of Narcissus papyraceus and Leucojum aestivum, cloned and expressed in Escherichia coli. Enzymatic assays performed with tyramine and 3,4-DHBA with each enzyme separately or combined, suggested that NBS and NR function together for the condensation of tyramine and 3,4-DHBA into norcraugsodine and further reduction into norbelladine. Using molecular homology modeling and docking studies, we predicted models for the binding of tyramine and 3,4-DHBA to NBS, and of the intermediate norcraugsodine to NR. Moreover, we show that NBS and NR physically interact in yeast and in-planta, that both localize to the cytoplasm and nucleus and are expressed at high levels in bulbs, confirming their colocalization and co-expression thus their ability to work together in the same catalytic route. Finally, their co-expression in yeast led to the production of norbelladine. In all, our study establishes that both NBS and NR participate in the biosynthesis of norbelladine by catalyzing the first key steps associated in the biosynthesis of the Alzheimer's drug galanthamine.
Keywords: Amaryllidaceae alkaloids; Leucojum aestivum; Narcissus papyraceus; enzyme activity; norbelladine synthase; noroxomaritidine/norcraugsodine reductase.
Copyright © 2023 Majhi, Gélinas, Mérindol, Ricard and Desgagné-Penix.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Battersby A. R., Fales H. M., Wildman W. C. (1961). Biosynthesis in the Amaryllidaceae. Tyrosine and norbelladine as precursors of haemanthamine. J. Amer. Chem. Soc 83, 4098–4099. doi: 10.1021/ja01480a037 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

