Changes in subcutaneous white adipose tissue cellular composition and molecular programs underlie glucose intolerance in persons with HIV
- PMID: 37711619
- PMCID: PMC10499182
- DOI: 10.3389/fimmu.2023.1152003
Changes in subcutaneous white adipose tissue cellular composition and molecular programs underlie glucose intolerance in persons with HIV
Abstract
Introduction: Subcutaneous adipose tissue (SAT) is a critical regulator of systemic metabolic homeostasis. Persons with HIV (PWH) have an increased risk of metabolic diseases and significant alterations in the SAT immune environment compared with the general population.
Methods: We generated a comprehensive single-cell multi-omic SAT atlas to characterize cellular compositional and transcriptional changes in 59 PWH across a spectrum of metabolic health.
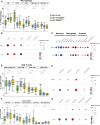
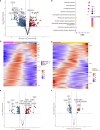
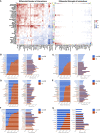
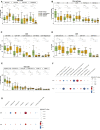
Results: Glucose intolerance was associated with increased lipid-associated macrophages, CD4+ and CD8+ T effector memory cells, and decreased perivascular macrophages. We observed a coordinated intercellular regulatory program which enriched for genes related to inflammation and lipid-processing across multiple cell types as glucose intolerance increased. Increased CD4+ effector memory tissue-resident cells most strongly associated with altered expression of adipocyte genes critical for lipid metabolism and cellular regulation. Intercellular communication analysis demonstrated enhanced pro-inflammatory and pro-fibrotic signaling between immune cells and stromal cells in PWH with glucose intolerance compared with non-diabetic PWH. Lastly, while cell type-specific gene expression among PWH with diabetes was globally similar to HIV-negative individuals with diabetes, we observed substantially divergent intercellular communication pathways.
Discussion: These findings suggest a central role of tissue-resident immune cells in regulating SAT inflammation among PWH with metabolic disease, and underscore unique mechanisms that may converge to promote metabolic disease.
Keywords: glucose intolerance; human immunodeficiency virus; immune cells; single-cell RNA sequencing; subcutaneous adipose tissue; type 2 diabetes mellitus; white adipose tissue.
Copyright © 2023 Bailin, Kropski, Gangula, Hannah, Simmons, Mashayekhi, Ye, Fan, Mallal, Warren, Kalams, Gabriel, Wanjalla and Koethe.
Conflict of interest statement
JKr consults for Boehringer Ingelheim, Janssen, and Bristol-Myers Squibb, serves on the scientific advisory board for APIE Therapeutics, and provides non-financial study support to Genentech. JKo has served as a consultant to Gilead Sciences, Merck, ViiV Healthcare, Theratechnologies, and Janssen, and has received research support from Gilead Sciences and Merck. CeW has served as a consultant for ViiV and has received research support from Gilead Sciences. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures





Similar articles
-
Adipose Tissue in Persons With HIV Is Enriched for CD4+ T Effector Memory and T Effector Memory RA+ Cells, Which Show Higher CD69 Expression and CD57, CX3CR1, GPR56 Co-expression With Increasing Glucose Intolerance.Front Immunol. 2019 Mar 19;10:408. doi: 10.3389/fimmu.2019.00408. eCollection 2019. Front Immunol. 2019. PMID: 30941121 Free PMC article.
-
Relationship of Subcutaneous Adipose Tissue Inflammation-Related Gene Expression With Ectopic Lipid Deposition in Persons With HIV.J Acquir Immune Defic Syndr. 2022 Jun 1;90(2):175-183. doi: 10.1097/QAI.0000000000002926. J Acquir Immune Defic Syndr. 2022. PMID: 35125474 Free PMC article.
-
A Distinct Form of Subcutaneous Fat Fibrosis Predicts Insulin Resistance in People with HIV.medRxiv [Preprint]. 2025 Jun 10:2025.05.13.25327547. doi: 10.1101/2025.05.13.25327547. medRxiv. 2025. PMID: 40463535 Free PMC article. Preprint.
-
Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance.Biochim Biophys Acta. 2014 Mar;1842(3):446-62. doi: 10.1016/j.bbadis.2013.05.017. Epub 2013 May 22. Biochim Biophys Acta. 2014. PMID: 23707515 Free PMC article. Review.
-
Adipose tissue regulates insulin sensitivity: role of adipogenesis, de novo lipogenesis and novel lipids.J Intern Med. 2016 Nov;280(5):465-475. doi: 10.1111/joim.12540. Epub 2016 Oct 3. J Intern Med. 2016. PMID: 27699898 Free PMC article. Review.
Cited by
-
Multiomic single-cell sequencing defines tissue-specific responses in Stevens-Johnson Syndrome and Toxic epidermal necrolysis.bioRxiv [Preprint]. 2024 Feb 18:2023.11.26.568771. doi: 10.1101/2023.11.26.568771. bioRxiv. 2024. Update in: Nat Commun. 2024 Oct 8;15(1):8722. doi: 10.1038/s41467-024-52990-3. PMID: 38405793 Free PMC article. Updated. Preprint.
-
Single-Cell Analysis of Subcutaneous Fat Reveals Profibrotic Cells That Correlate With Visceral Adiposity in HIV.J Clin Endocrinol Metab. 2024 Dec 18;110(1):238-253. doi: 10.1210/clinem/dgae369. J Clin Endocrinol Metab. 2024. PMID: 38820087 Free PMC article.
-
The Primacy of Adipose Tissue Gene Expression and Plasma Lipidome in Cardiometabolic Disease in Persons With HIV.J Infect Dis. 2025 Feb 20;231(2):e407-e418. doi: 10.1093/infdis/jiae532. J Infect Dis. 2025. PMID: 39657693 Free PMC article.
-
Subcutaneous adipose tissue gene transcripts associated with progressive myosteatosis in persons with HIV.AIDS. 2025 Oct 1;39(12):1739-1748. doi: 10.1097/QAD.0000000000004264. Epub 2025 Jun 11. AIDS. 2025. PMID: 40528404
-
The pathogenesis of obesity in people living with HIV.Curr Opin HIV AIDS. 2024 Jan 1;19(1):6-13. doi: 10.1097/COH.0000000000000834. Epub 2023 Nov 6. Curr Opin HIV AIDS. 2024. PMID: 37934696 Free PMC article. Review.
References
-
- Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. . Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract (2019) 157:107843. doi: 10.1016/j.diabres.2019.107843 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR024975/RR/NCRR NIH HHS/United States
- T32 AI007474/AI/NIAID NIH HHS/United States
- R01 DK131529/DK/NIDDK NIH HHS/United States
- R25 HL145817/HL/NHLBI NIH HHS/United States
- K23 DK135414/DK/NIDDK NIH HHS/United States
- T32 HL007444/HL/NHLBI NIH HHS/United States
- K23 HL156759/HL/NHLBI NIH HHS/United States
- R01 HL145372/HL/NHLBI NIH HHS/United States
- KL2 TR002245/TR/NCATS NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- K12 HL143956/HL/NHLBI NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- R01 DK112262/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

