Single-nucleobase resolution of a surface energy transfer nanoruler for in situ measurement of aptamer binding at the receptor subunit level in living cells
- PMID: 37712043
- PMCID: PMC10498721
- DOI: 10.1039/d3sc01244a
Single-nucleobase resolution of a surface energy transfer nanoruler for in situ measurement of aptamer binding at the receptor subunit level in living cells
Abstract
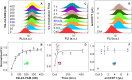
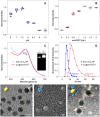
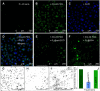
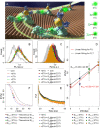
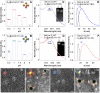
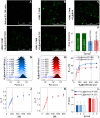
In situ identification of aptamer-binding targets on living cell membrane surfaces is of considerable interest, but a major challenge, specifically, when advancing recognition to the level of membrane receptor subunits. Here we propose a novel nanometal surface energy transfer (NSET) based nanoruler with a single-nucleobase resolution (SN-nanoruler), in which FAM-labeled aptamers and single-sized gold nanoparticle (GNP) antibody conjugates act as a donor and an acceptor. A single nucleobase resolution of the SN-nanoruler was experimentally illustrated by molecular size, orientation, quenching nature, and other dye-GNP pairs. The SN-nanoruler provides high reproducibility and precision for measuring molecule distance on living cell membranes at the nanometer level owing to only the use of single-sized antibody-capped GNPs. In situ identification of the aptamer binding site was advanced to the protein subunit level on the living cell membrane for the utilization of this SN-nanoruler. The results suggest that the proposed strategy is a solid step towards the wider application of optical-based rulers to observe the molecular structural configuration and dynamic transitions on the membrane surface of living cells.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Single-Nucleobase-Resolved Nanoruler Determines the Surface Energy Transfer Radius on the Living Cell Membrane.Anal Chem. 2024 Apr 2;96(13):5274-5281. doi: 10.1021/acs.analchem.4c00128. Epub 2024 Mar 20. Anal Chem. 2024. PMID: 38507515
-
Record Resolution of Nanometal Surface Energy Transfer Optical Nanoruler Projects 3D Spatial Configuration of Aptamers on a Living Cell Membrane.Nano Lett. 2023 Dec 27;23(24):11968-11974. doi: 10.1021/acs.nanolett.3c04322. Epub 2023 Dec 7. Nano Lett. 2023. PMID: 38059895
-
Plasmon Resonance Energy Transfer Nanoruler for Pinpointing Molecular Distance and Interaction on the Living Cell Membrane.Nano Lett. 2023 Aug 23;23(16):7750-7757. doi: 10.1021/acs.nanolett.3c01629. Epub 2023 Jun 30. Nano Lett. 2023. PMID: 37387534
-
A surface energy transfer nanoruler for measuring binding site distances on live cell surfaces.J Am Chem Soc. 2010 Nov 24;132(46):16559-70. doi: 10.1021/ja106360v. Epub 2010 Nov 1. J Am Chem Soc. 2010. PMID: 21038856 Free PMC article.
-
Recent Advances in Aptamer-Based Nanopore Sensing at Single-Molecule Resolution.Chem Asian J. 2022 Aug 15;17(16):e202200364. doi: 10.1002/asia.202200364. Epub 2022 Jun 23. Chem Asian J. 2022. PMID: 35644914 Review.
Cited by
-
Transmembrane nanogold energy transfer ruler enables in situ synchronized projecting of receptor dimerization-phosphorylation signaling.Chem Sci. 2025 Jul 29. doi: 10.1039/d5sc02717a. Online ahead of print. Chem Sci. 2025. PMID: 40740738 Free PMC article.
References
-
- Shangguan D. Li Y. Tang Z. Cao Z. C. Chen H. W. Mallikaratchy P. Sefah K. Yang C. J. Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U.S.A. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. doi: 10.1073/pnas.0602615103. - DOI - DOI - PMC - PubMed
-
- Miao X. Li Z. Zhu A. Feng Z. Tian J. Peng X. Ultrasensitive electrochemical detection of protein tyrosine kinase-7 by gold nanoparticles and methylene blue assisted signal amplification. Biosens. Bioelectron. 2016;83:39–44. doi: 10.1016/j.bios.2016.04.032. doi: 10.1016/j.bios.2016.04.032. - DOI - DOI - PubMed
LinkOut - more resources
Full Text Sources

