Neuromusculoskeletal model-informed machine learning-based control of a knee exoskeleton with uncertainties quantification
- PMID: 37712095
- PMCID: PMC10498472
- DOI: 10.3389/fnins.2023.1254088
Neuromusculoskeletal model-informed machine learning-based control of a knee exoskeleton with uncertainties quantification
Abstract
Introduction: Research interest in exoskeleton assistance strategies that incorporate the user's torque capacity is growing rapidly. However, the predicted torque capacity from users often includes uncertainty from various sources, which can have a significant impact on the safety of the exoskeleton-user interface.
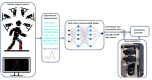
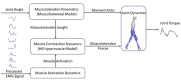
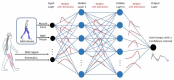
Methods: To address this challenge, this paper proposes an adaptive control framework for a knee exoskeleton that uses muscle electromyography (EMG) signals and joint kinematics. The framework predicted the user's knee flexion/extension torque with confidence bounds to quantify the uncertainty based on a neuromusculoskeletal (NMS) solver-informed Bayesian Neural Network (NMS-BNN). The predicted torque, with a specified confidence level, controlled the assistive torque provided by the exoskeleton through a TCP/IP stream. The performance of the NMS-BNN model was also compared to that of the Gaussian process (NMS-GP) model.
Results: Our findings showed that both the NMS-BNN and NMS-GP models accurately predicted knee joint torque with low error, surpassing traditional NMS models. High uncertainties were observed at the beginning of each movement, and at terminal stance and terminal swing in self-selected speed walking in both NMS-BNN and NMS-GP models. The knee exoskeleton provided the desired assistive torque with a low error, although lower torque was observed during terminal stance of fast walking compared to self-selected walking speed.
Discussion: The framework developed in this study was able to predict knee flexion/extension torque with quantifiable uncertainty and to provide adaptive assistive torque to the user. This holds significant potential for the development of exoskeletons that provide assistance as needed, with a focus on the safety of the exoskeleton-user interface.
Keywords: data-driven biomechanical models; inverse dynamics; machine learning; neuromusculoskeletal modeling; uncertainty quantification.
Copyright © 2023 Zhang, Zhang, Zhu, Wang and Gutierrez-Farewik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Estimation of Joint Torque by EMG-Driven Neuromusculoskeletal Models and LSTM Networks.IEEE Trans Neural Syst Rehabil Eng. 2023;31:3722-3731. doi: 10.1109/TNSRE.2023.3315373. Epub 2023 Sep 22. IEEE Trans Neural Syst Rehabil Eng. 2023. PMID: 37708013
-
Ankle Joint Torque Prediction Using an NMS Solver Informed-ANN Model and Transfer Learning.IEEE J Biomed Health Inform. 2022 Dec;26(12):5895-5906. doi: 10.1109/JBHI.2022.3207313. Epub 2022 Dec 7. IEEE J Biomed Health Inform. 2022. PMID: 36112547
-
Heuristic-Based Ankle Exoskeleton Control for Co-Adaptive Assistance of Human Locomotion.IEEE Trans Neural Syst Rehabil Eng. 2019 Oct;27(10):2059-2069. doi: 10.1109/TNSRE.2019.2936383. Epub 2019 Aug 19. IEEE Trans Neural Syst Rehabil Eng. 2019. PMID: 31425120
-
Human-in-the-Loop Optimization of Knee Exoskeleton Assistance for Minimizing User's Metabolic and Muscular Effort.Sensors (Basel). 2024 May 22;24(11):3305. doi: 10.3390/s24113305. Sensors (Basel). 2024. PMID: 38894101 Free PMC article.
-
Exoskeletons: A challenge for development.Wearable Technol. 2023 Jan 5;4:e1. doi: 10.1017/wtc.2022.28. eCollection 2023. Wearable Technol. 2023. PMID: 38487778 Free PMC article. Review.
Cited by
-
Soft ankle exoskeleton to counteract dropfoot and excessive inversion.Front Neurorobot. 2024 Aug 21;18:1372763. doi: 10.3389/fnbot.2024.1372763. eCollection 2024. Front Neurorobot. 2024. PMID: 39234442 Free PMC article.
References
-
- Bergstra J., Bengio Y. (2012). Random search for hyper-parameter optimization. J. Mach. Learn. Res. 13, 1–25.
-
- Cao H., Chen G., Li Z., Feng Q., Lin J., Knoll A. (2022a). Efficient grasp detection network with Gaussian-based grasp representation for robotic manipulation. IEEE/ASME Trans. Mechatron. 28, 1384–1394. 10.1109/TMECH.2022.3224314 - DOI
-
- Cao H., Chen G., Li Z., Hu Y., Knoll A. (2022b). Neurograsp: multimodal neural network with Euler region regression for neuromorphic vision-based grasp pose estimation. IEEE Trans. Instrument. Meas. 71, 1–11. 10.1109/TIM.2022.3179469 - DOI
LinkOut - more resources
Full Text Sources

